|
|
|
|
GEO 365N/384S Seismic Data Processing Computational Assignment 3 |
In this part of the assignment, we will apply NMO-based velocity analysis to the Viking Graben dataset (Keys and Foster, 1998). The input data for this assignment have been preprocessed by applying multiple attenuation using the parabolic Radon transform (we will discuss this method later in the class).
scons -cto remove (clean) previously generated files.
scons cmps.viewto display the input data (Figure 1.)

|
|---|
|
cmps
Figure 1. Viking Graben dataset after preprocessing and sorting into CMP gathers. |
|
|
scons cmp.viewto display the selected gather (Figure 2.)

|
|---|
|
cmp
Figure 2. CMP gather selected for velocity analysis. |
|
|
Run
scons nmo1.viewto observe the result of velocity analysis on the selected CMP gather using automatic picking (Figure 3a.)
Questions: Why does seismic velocity usually increase with depth? What can be the physical nature of low-velocity seismic events appearing at later times?

To guide the automatic picker to higher velocities, we can apply a muting function to the semblance gather. Simple muting by cutting a lower-left triangle corner from the semblance scan is implemented in the attached mute.c function.
Run
scons nmo2.viewto observe the effect of muting (Figure 3a.)


|
|---|
|
nmo1,nmo2
Figure 3. NMO velocity analysis with automatic picking applied to the selected CMP gather: (a) Initial pick, (b) Pick guided by muting. |
|
|
Run
scons vstack.viewto run the semblance analysis with automatic picking on every CMP gather (this may take a while depending on the speed and the number of cores of your computer). The result is shown in Figure 4.

|
|---|
|
vstack
Figure 4. NMO stacking velocity picked automatically from the whole line. |
|
|
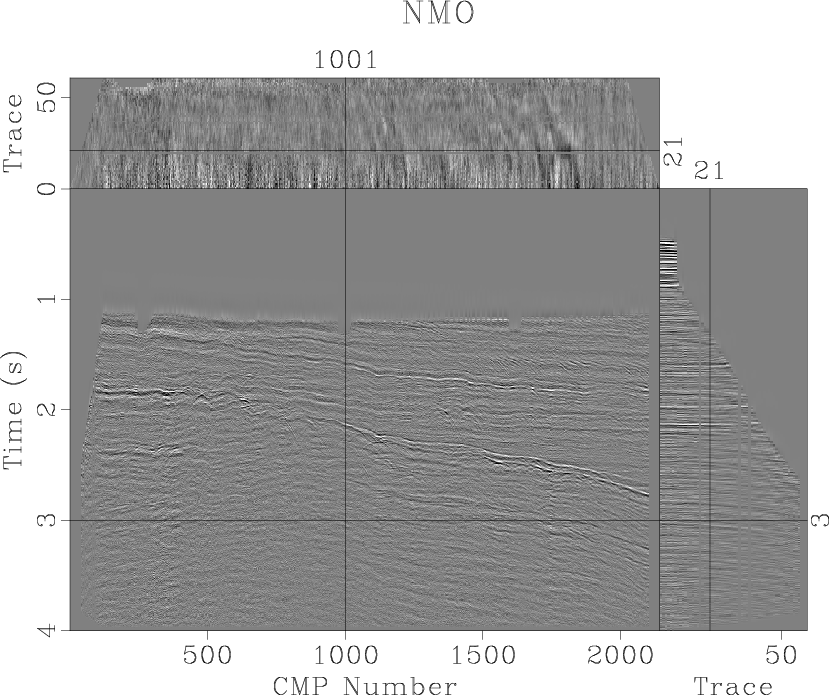
Run
scons nmo.view scons stack.viewto display the result (Figures 5 and 6.)
|
|---|
|
nmo
Figure 5. Viking Graben dataset after the normal-moveout correction. |
|
|

|
|---|
|
stack
Figure 6. NMO stack of the Viking Graben dataset. |
|
|
scons nmo.vpl
The last section of the SConstruct file generates velocity analysis plots for several selected gathers (Figure 7.)

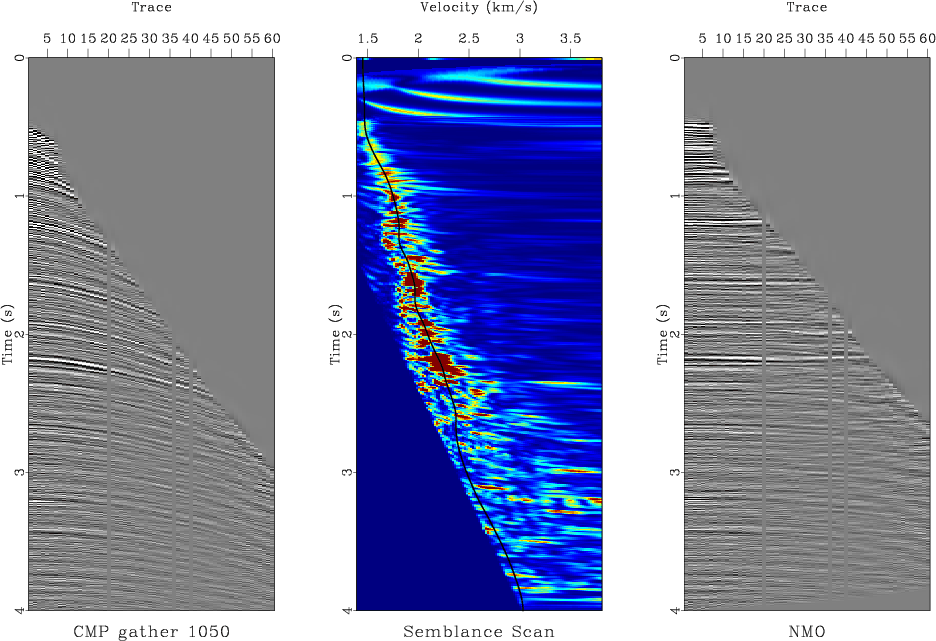


|
|---|
|
cmp700,cmp1050,cmp1400,cmp1750
Figure 7. NMO velocity analysis with automatic picking displayed for selected CMP gathers. |
|
|
Describe the method you used to improve the results:

/* Mute a triangle region */
#include <rsf.h>
int main(int argc, char* argv[])
{
int n1, n2, n3, i1, i2, i3;
float t0,v0, dt,dv, t1, v1, slope,tmax,band, t,taper, *data;
sf_file in, out;
sf_init (argc,argv);
in = sf_input("in");
out = sf_output("out");
if (!sf_histint(in,"n1",&n1)) sf_error("No n1= in input");
if (!sf_histint(in,"n2",&n2)) sf_error("No n2= in input");
n3 = sf_leftsize(in,2);
if (!sf_histfloat(in,"o1",&t0)) sf_error("No o1= in input");
if (!sf_histfloat(in,"d1",&dt)) sf_error("No d1= in input");
if (!sf_histfloat(in,"o2",&v0)) sf_error("No o2= in input");
if (!sf_histfloat(in,"d2",&dv)) sf_error("No d2= in input");
if (!sf_getfloat("t1",&t1)) t1=t0;
/* start time */
if (!sf_getfloat("v1",&v1)) v1=v0+(n2-1)*dv;
/* end velocity */
if (!sf_getfloat("band",&band)) band=20*dt;
/* start time */
slope = (t0+(n1-1)*dt-t1)/(v1-v0);
data = sf_floatalloc(n1);
for (i3=0; i3 < n3; i3++) {
for (i2=0; i2 < n2; i2++) {
tmax = t1 + i2*dv*slope;
sf_floatread (data,n1,in);
for (i1=0; i1 < n1; i1++) {
t = t0+i1*dt;
if (t > tmax) {
data[i1] = 0.0f;
} else if (t > tmax-band) {
taper = sinf(0.5 * SF_PI * (t-tmax)/band);
data[i1] *= taper*taper;
}
}
sf_floatwrite (data,n1,out);
}
}
exit(0);
}
|
from rsf.proj import *
# Download pre-processed CMP gathers
# from the Viking Graben dataset
Fetch('paracdp.segy','viking')
# Convert to RSF
Flow('paracdp tparacdp','paracdp.segy',
'segyread tfile=${TARGETS[1]}')
# Convert to CDP gathers, time-power gain and high-pass filter
Flow('cmps','paracdp',
'''
intbin xk=cdpt yk=cdp | window max1=4 |
pow pow1=2 | bandpass flo=5
''')
Result('cmps',
'''
byte gainpanel=all | transp plane=23 memsize=5000 |
grey3 frame1=750 frame2=1000 frame3=20
point1=0.8 point2=0.8
title="CMP Gathers" label2="CMP Number"
''')
# Extract offsets
Flow('offsets mask','tparacdp',
'''
headermath output=offset |
intbin head=$SOURCE xk=cdpt yk=cdp mask=${TARGETS[1]} |
dd type=float |
scale dscale=0.001
''')
# Window bad traces
Flow('maskbad','cmps',
'''
mul $SOURCE | stack axis=1 | cut n1=1 n2=1 f2=712 |
mask min=1e-20
''')
Flow('mask2','maskbad mask','spray axis=1 n=1 | mul ${SOURCES[1]}')
# Extract one CMP gather
########################
Flow('mask1','mask2','window n3=1 f3=1000 squeeze=n')
Flow('cmp','cmps mask1',
'window n3=1 f3=1000 | headerwindow mask=${SOURCES[1]}')
Flow('offset','offsets mask1',
'''
window n3=1 f3=1000 squeeze=n |
headerwindow mask=${SOURCES[1]}
''')
Result('cmp','cmp offset',
'''
wiggle poly=y yreverse=y transp=y xpos=${SOURCES[1]}
label2=Offset unit2=km title="CMP Gather"
''')
Plot('cmp','grey title="CMP gather"')
# Velocity scan
Flow('vscan1','cmp offset',
'''
vscan semblance=y half=n v0=1.4 nv=121 dv=0.02
offset=${SOURCES[1]}
''')
Plot('vscan1',
'grey color=j allpos=y title="Semblance Scan" unit2=km/s')
# Automatic pick
Flow('vpick1','vscan1','pick rect1=25 vel0=1.45')
Plot('vpick1',
'''
graph yreverse=y transp=y plotcol=7 plotfat=7
pad=n min2=1.4 max2=3.8 wantaxis=n wanttitle=n
''')
Plot('vscan2','vscan1 vpick1','Overlay')
# NMO
Flow('nmo1','cmp offset vpick1',
'nmo half=n offset=${SOURCES[1]} velocity=${SOURCES[2]}')
Plot('nmo1','grey title=NMO')
Result('nmo1','cmp vscan2 nmo1','SideBySideAniso',
vppen='txscale=1.5')
mute = Program('mute.c')
Flow('vmute','vscan1 %s' % mute[0],'./${SOURCES[1]} t1=1.5 v1=3')
Plot('vmute',
'grey color=j allpos=y title="Semblance Scan" unit2=km/s')
Flow('vpick2','vmute','pick rect1=25 vel0=1.45')
Plot('vpick2',
'''
graph yreverse=y transp=y plotcol=7 plotfat=7
pad=n min2=1.4 max2=3.8 wantaxis=n wanttitle=n
''')
Plot('vmute2','vmute vpick2','Overlay')
Flow('nmo2','cmp offset vpick2',
'nmo half=n offset=${SOURCES[1]} velocity=${SOURCES[2]}')
Plot('nmo2','grey title=NMO')
Result('nmo2','cmp vmute2 nmo2','SideBySideAniso',
vppen='txscale=1.5')
# Apply to all CMP gathers
##########################
Flow('vscan','cmps offsets mask2 %s' % mute[0],
'''
vscan semblance=y half=n v0=1.4 nv=121 dv=0.02
offset=${SOURCES[1]} mask=${SOURCES[2]} |
./${SOURCES[3]} t1=1.5 v1=3
''',
split=[3,'omp'])
Flow('vpick','vscan','pick rect1=25 rect2=50 vel0=1.45')
# Automatic pick
Flow('vstack','vpick','transp | remap1 n1=2142 d1=1 o1=1 | transp')
Result('vstack',
'''
grey mean=y color=j scalebar=y barreverse=y barunit=km/s
title="Picked Stacking Velocity" label2="CMP Number" unit2=
''')
# Apply NMO and stack
Flow('vnmo','vstack','spray axis=2 n=1')
Flow('nmo','cmps offsets mask2 vnmo',
'''
nmo half=n offset=${SOURCES[1]}
mask=${SOURCES[2]} velocity=${SOURCES[3]}
''',split=[3,'omp'])
Result('nmo',
'''
byte gainpanel=all | transp plane=23 memsize=5000 |
grey3 frame1=750 frame2=1000 frame3=20
title=NMO label2="CMP Number" point1=0.8 point2=0.8
''')
Plot('nmo',
'''
window j3=10 | byte gainpanel=all |
grey3 frame1=500 frame2=20 movie=3
title=NMO label3="CMP Number"
''',view=1)
Flow('stack','nmo','stack')
Result('stack',
'grey title="Viking Graben Stack" label2="CMP Number" ')
# Automatic gain control
Flow('agc','stack','shapeagc rect1=250')
Result('agc','grey title="AGC Stack" label2="CMP Number" ')
# Examine NMO on individual gathers
###################################
for p in (350,700,1050,1400,1750):
cdp = 'cmp%d' % p
Flow(cdp,'cmps','window n3=1 f3=%d' % p)
Plot(cdp,'grey title="CMP gather %d"' % p)
vscan = 'vscan%d' % p
Flow(vscan,'vscan','window n3=1 f3=%d' % p)
Plot(vscan,
'grey color=j allpos=y title="Semblance Scan" unit2=km/s')
vpick = 'vpick%d' % p
Flow(vpick,'vstack','window n2=1 f2=%d' % p)
Plot(vpick,
'''
graph yreverse=y transp=y plotcol=7 plotfat=7
pad=n min2=1.4 max2=3.8 wantaxis=n wanttitle=n
''')
Plot(vscan+'2',[vscan,vpick],'Overlay')
nmo = 'nmo%d' % p
Flow(nmo,'nmo','window n3=1 f3=%d' % p)
Plot(nmo,'grey title=NMO')
Result(cdp,[cdp,vscan+'2',nmo],'SideBySideAniso',
vppen='txscale=1.5')
End()
|
|
|
|
|
GEO 365N/384S Seismic Data Processing Computational Assignment 3 |