|
|
|
|
GEO 365N/384S Seismic Data Processing Computational Assignment 5 |
The parabolic Radon transform is a powerful tool but not perfectly suitable for separating hyperbolic events. An alternative tool is the time-domain hyperbolic Radon transform (also known as the velocity stack or the velocity transform). The idea of making the hyperbolic Radon transform invertible by iterative least-squares inversion was proposed by Thorson and Claerbout (1985).
scons -cto remove (clean) previously generated files.
|
|---|
|
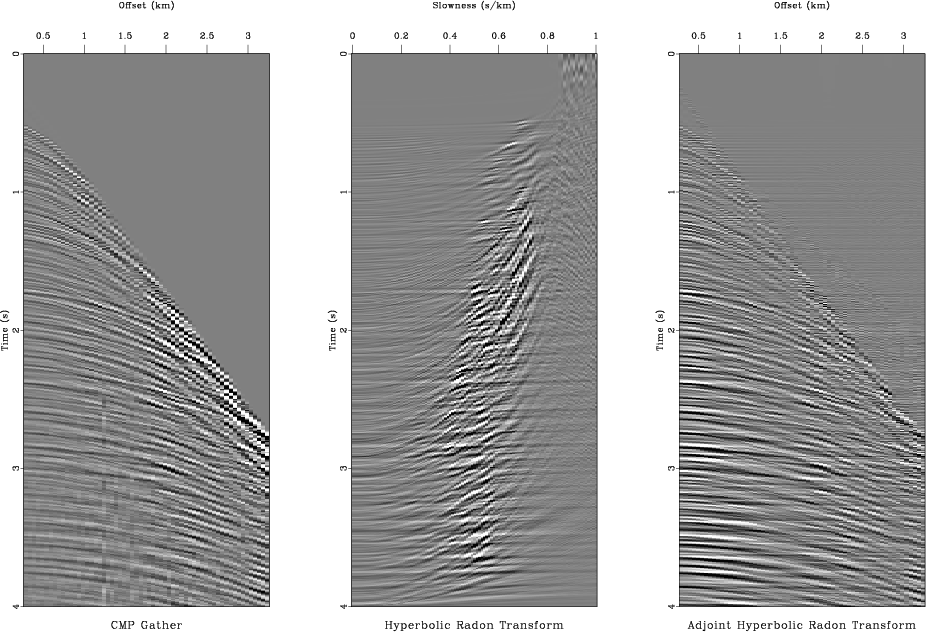
adj
Figure 8. Applying the hyperbolic Radon transform. From left to right: selected CMP gather, its hyperbolic Radon transform, data reconstructed by the adjoint transform (filtered back projection). |
|
|
scons adj.view

sfdottest ./hradon.exe np=101 dp=0.01 op=0 nx=60 dx=0.05 ox=0.287 \ mod=rad.rsf dat=cmp.rsfThe output should display a pair of numbers equal to six digits of accurate. You can run the test multiple times.

|
|---|
|
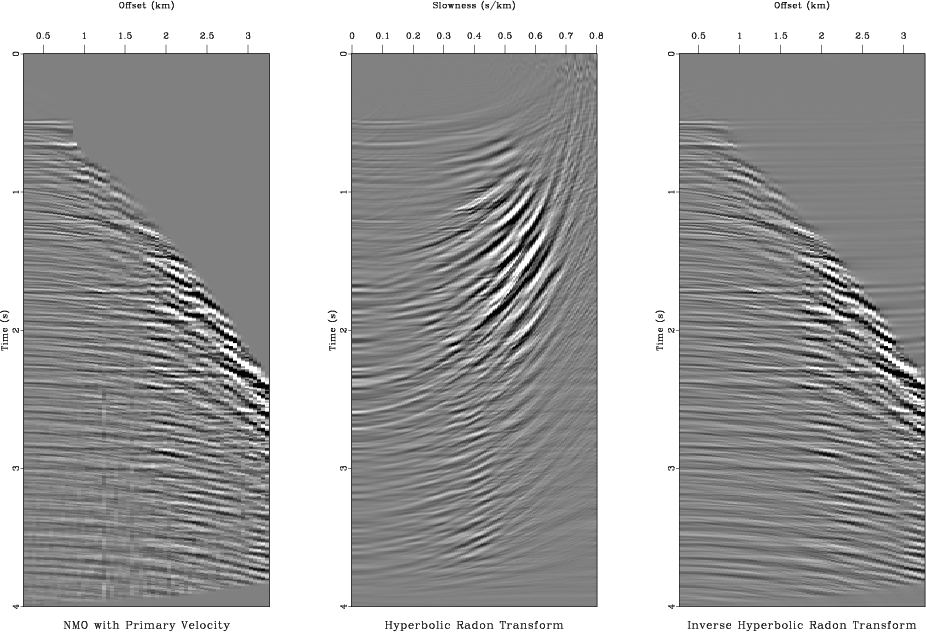
inv
Figure 9. Applying the hyperbolic Radon transform. From left to right: selected CMP gather, its hyperbolic Radon transform (using iterative least-squares inversion), data reconstructed by the inverse transform. |
|
|
scons inv.view
|
|---|
|
nmorad
Figure 10. Applying the hyperbolic Radon transform after NMO with the primary velocity trend. From left to right: selected CMP gather after NMO, hyperbolic Radon transform (using iterative least-squares inversion), data reconstructed by the inverse transform. |
|
|
|
|---|
|
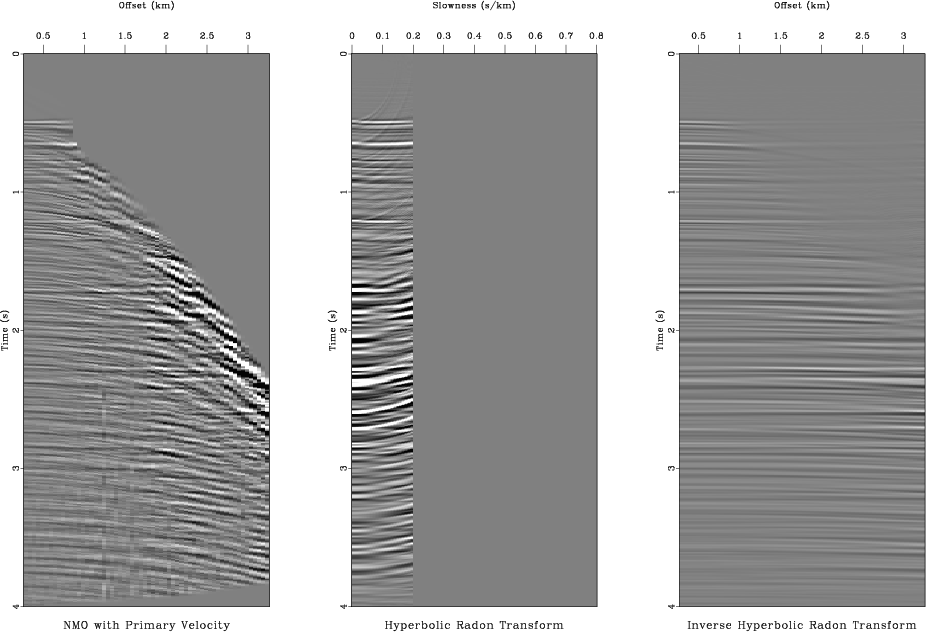
nmocut
Figure 11. Isolating signal in the hyperbolic Radon domain. From left to right: hyperbolic Radon transform, isolated nearly-flat events, signal reconstructed by the inverse transform. |
|
|
|
|---|
|
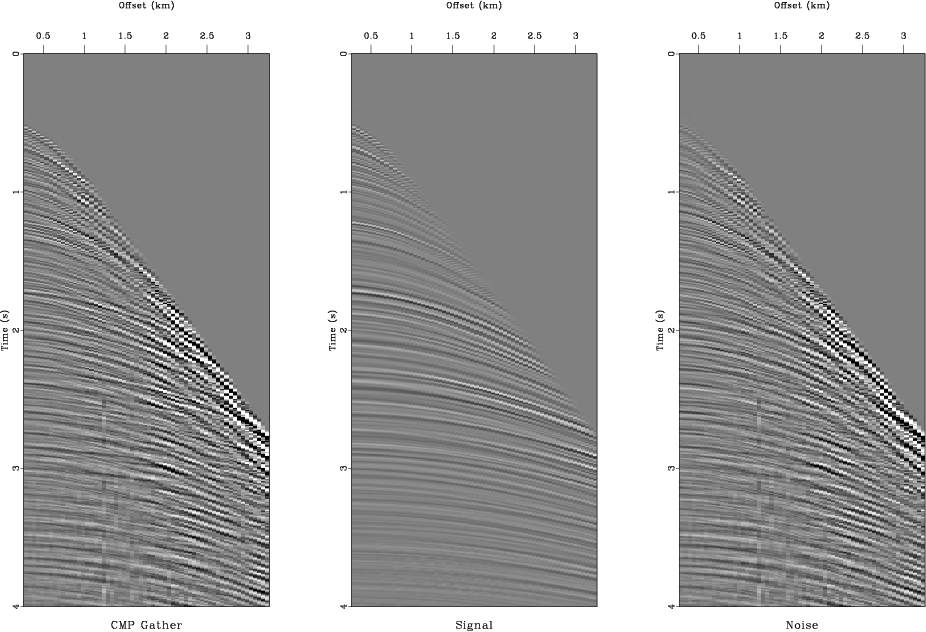
inmo,pvscan
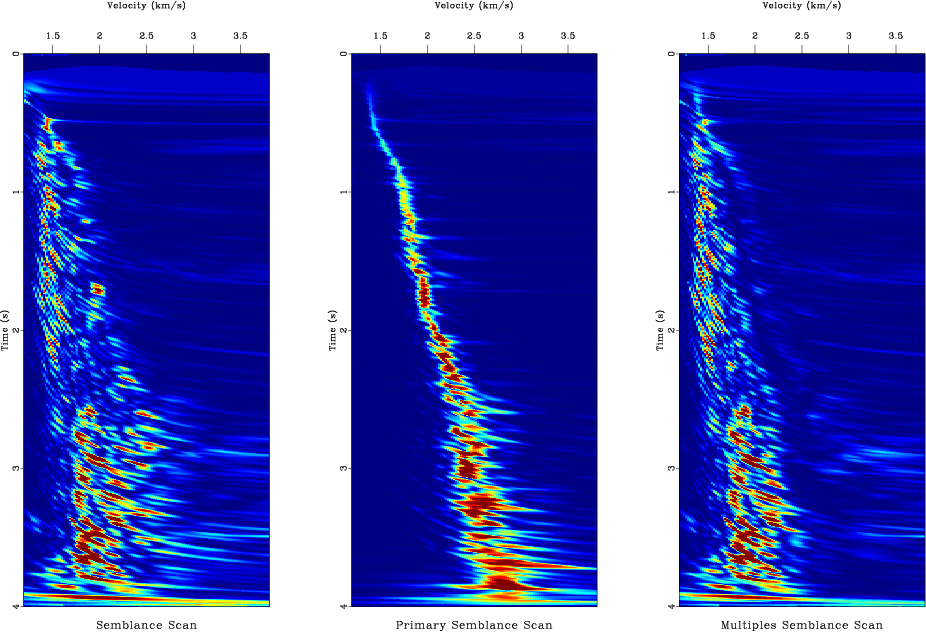
Figure 12. Separating signal (primary reflections) and noise (multiple reflections). (a) From left to right: input CMP gather, estimated primaries, estimated multiples. (b) Velocity analysis using semblance scan. From left to right: using all data, using estimated primaries, using estimated multiples. |
|
|
The hyperbolic Radon transform is shown in Figure 10 and the output of the procedure described above is shown in Figure 12. To reproduce them on your screen, run
scons inmo.view pvscan.view
/* Hyperbolic Radon transform */
#include <rsf.h>
int main(int argc, char* argv[])
{
sf_map4 map;
int it,nt, ix,nx, ip,np, i3,n3;
bool adj;
float t0,dt,t, x0,dx,x, p0,dp,p;
float **cmp, **rad, *trace;
sf_file in, out;
sf_init(argc,argv);
in = sf_input("in");
out = sf_output("out");
if (!sf_histint(in,"n1",&nt)) sf_error("No n1=");
if (!sf_histfloat(in,"o1",&t0)) sf_error("No o1=");
if (!sf_histfloat(in,"d1",&dt)) sf_error("No d1=");
n3 = sf_leftsize(in,2);
if (!sf_getbool("adj",&adj)) adj=false;
/* adjoint flag */
if (adj) {
if (!sf_histint(in,"n2",&nx)) sf_error("No n2=");
if (!sf_histfloat(in,"o2",&x0)) sf_error("No o2=");
if (!sf_histfloat(in,"d2",&dx)) sf_error("No d2=");
if (!sf_getint("np",&np)) sf_error("Need np=");
if (!sf_getfloat("op",&p0)) sf_error("need p0=");
if (!sf_getfloat("dp",&dp)) sf_error("need dp=");
sf_putint(out,"n2",np);
sf_putfloat(out,"o2",p0);
sf_putfloat(out,"d2",dp);
} else {
if (!sf_histint(in,"n2",&np)) sf_error("No n2=");
if (!sf_histfloat(in,"o2",&p0)) sf_error("No o2=");
if (!sf_histfloat(in,"d2",&dp)) sf_error("No d2=");
if (!sf_getint("nx",&nx)) sf_error("Need nx=");
if (!sf_getfloat("ox",&x0)) sf_error("need x0=");
if (!sf_getfloat("dx",&dx)) sf_error("need dx=");
sf_putint(out,"n2",nx);
sf_putfloat(out,"o2",x0);
sf_putfloat(out,"d2",dx);
}
trace = sf_floatalloc(nt);
cmp = sf_floatalloc2(nt,nx);
rad = sf_floatalloc2(nt,np);
/* initialize half-order differentiation */
sf_halfint_init (true,nt,1.0f-1.0f/nt);
/* initialize spline interpolation */
map = sf_stretch4_init (nt, t0, dt, nt, 0.01);
for (i3=0; i3 < n3; i3++) {
if( adj) {
for (ix=0; ix < nx; ix++) {
sf_floatread(trace,nt,in);
sf_halfint_lop(true,false,nt,nt,cmp[ix],trace);
}
} else {
sf_floatread(rad[0],nt*np,in);
}
sf_adjnull(adj,false,nt*np,nt*nx,rad[0],cmp[0]);
for (ip=0; ip < np; ip++) {
p = p0 + ip*dp;
for (ix=0; ix < nx; ix++) {
x = x0 + ix*dx;
for (it=0; it < nt; it++) {
t = t0 + it*dt;
trace[it] = hypotf(t,p*x);
/* hypot(a,b)=sqrt(a*a+b*b) */
}
sf_stretch4_define(map,trace);
if (adj) {
sf_stretch4_apply_adj(true,map,rad[ip],cmp[ix]);
} else {
sf_stretch4_apply (true,map,rad[ip],cmp[ix]);
}
}
}
if( adj) {
sf_floatwrite(rad[0],nt*np,out);
} else {
for (ix=0; ix < nx; ix++) {
sf_halfint_lop(false,false,nt,nt,cmp[ix],trace);
sf_floatwrite(trace,nt,out);
}
}
}
exit(0);
}
|
from rsf.proj import *
# Download one CMP gather (with mute)
Fetch('cmp.HH','viking')
Flow('cmp','cmp.HH','dd form=native')
Plot('cmp','grey title="CMP Gather" clip=8.66')
prog = Program('hradon.c')
hradon = str(prog[0])
Flow('rad',['cmp',hradon],
'./${SOURCES[1]} adj=y np=101 dp=0.01 op=0')
Plot('rad',
'''
grey title="Hyperbolic Radon Transform"
label2=Slowness unit2=s/km
''')
Flow('adj',['rad',hradon],
'./${SOURCES[1]} adj=n nx=60 dx=0.05 ox=0.287')
Plot('adj','grey title="Adjoint Hyperbolic Radon Transform" ')
Result('adj','cmp rad adj','SideBySideAniso')
Flow('inv',['cmp',hradon,'rad'],
'''
conjgrad ./${SOURCES[1]} mod=${SOURCES[2]}
np=101 dp=0.01 op=0 nx=60 dx=0.05 ox=0.287 niter=10
''')
Plot('inv',
'''
grey title="Hyperbolic Radon Transform"
label2=Slowness unit2=s/km
''')
Flow('cmp2',['inv',hradon],
'./${SOURCES[1]} adj=n nx=60 dx=0.05 ox=0.287')
Plot('cmp2',
'grey title="Inverse Hyperbolic Radon Transform" clip=8.66')
Result('inv','cmp inv cmp2','SideBySideAniso')
# Velocity scan
Flow('vscan','cmp',
'vscan semblance=y half=n v0=1.2 nv=131 dv=0.02')
Plot('vscan',
'grey color=j allpos=y title="Semblance Scan" unit2=km/s')
# Automatic pick
Flow('vpick','vscan',
'''
mutter inner=y x0=1.4 half=n v0=0.45 t0=0.5 |
pick rect1=50 vel0=1.5
''')
Plot('vpick',
'''
graph yreverse=y transp=y plotcol=7 plotfat=7
pad=n min2=1.2 max2=3.8 wantaxis=n wanttitle=n
''')
Plot('vscan2','vscan vpick','Overlay')
# NMO
Flow('nmo','cmp vpick','nmo half=n velocity=${SOURCES[1]}')
Plot('nmo','grey title="NMO with Primary Velocity" clip=9.83')
Result('vscan','cmp vscan2 nmo','SideBySideAniso')
Flow('nmorad0',['cmp',hradon],
'./${SOURCES[1]} adj=y np=161 dp=0.005 op=0')
Flow('nmorad',['nmo',hradon,'nmorad0'],
'''
conjgrad ./${SOURCES[1]} mod=${SOURCES[2]}
np=161 dp=0.005 op=0 nx=60 dx=0.05 ox=0.287 niter=10
''')
Plot('nmorad',
'''
grey title="Hyperbolic Radon Transform"
label2=Slowness unit2=s/km
''')
Flow('nmo2',['nmorad',hradon],
'./${SOURCES[1]} adj=n nx=60 dx=0.05 ox=0.287')
Plot('nmo2',
'grey title="Inverse Hyperbolic Radon Transform" clip=9.83')
Result('nmorad','nmo nmorad nmo2','SideBySideAniso')
Flow('cut','nmorad','cut min2=0.2')
Plot('cut',
'''
grey title="Hyperbolic Radon Transform"
label2=Slowness unit2=s/km
''')
Flow('signal',['cut',hradon],
'./${SOURCES[1]} adj=n nx=60 dx=0.05 ox=0.287')
Plot('signal',
'grey title="Inverse Hyperbolic Radon Transform" clip=9.83')
Result('nmocut','nmo cut signal','SideBySideAniso')
# Inverse NMO
Flow('inmo','signal vpick',
'inmo half=n velocity=${SOURCES[1]} | mutter half=n v0=1.2')
Plot('inmo','grey title=Signal clip=8.66')
Flow('pvscan','inmo',
'vscan semblance=y half=n v0=1.2 nv=131 dv=0.02')
Plot('pvscan',
'grey color=j allpos=y title="Primary Semblance Scan" ')
Flow('mult','cmp inmo','add scale=1,-1 ${SOURCES[1]}')
Plot('mult','grey title=Noise clip=8.66')
Flow('mvscan','mult',
'vscan semblance=y half=n v0=1.2 nv=131 dv=0.02')
Plot('mvscan',
'grey color=j allpos=y title="Multiples Semblance Scan" ')
Result('inmo','cmp inmo mult','SideBySideAniso')
Result('pvscan','vscan pvscan mvscan','SideBySideAniso')
End()
|
|
|
|
|
GEO 365N/384S Seismic Data Processing Computational Assignment 5 |