Guide to RSF file format: Difference between revisions
typos |
|||
| (37 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
<center><font size="-1">''This page was created from the LaTeX source in [ | <center><font size="-1">''This page was created from the LaTeX source in [https://github.com/ahay/src/blob/master/book/rsf/rsf/format.tex book/rsf/rsf/format.tex] using [[latex2wiki]]''</font></center> | ||
[[Image:Fotolia_9592362_XS.jpg|right|]] | |||
==Principles== | ==Principles== | ||
The main design principle behind the RSF | The main design principle behind the RSF data format is [http://en.wikipedia.org/wiki/KISS_principle KISS] ("Keep It | ||
Simple | Short and Simple"). The RSF format is borrowed from the SEPlib data format | ||
initially designed at the Stanford Exploration Project | |||
(Claerbout, 1991<ref>Claerbout, J. F., 1991, Introduction to Seplib and SEP utility software, ''in'' SEP-70, 413--436. Stanford Exploration Project.</ref>). The format is made as simple as possible for | (Claerbout, 1991<ref>Claerbout, J. F., 1991, Introduction to Seplib and SEP utility software, ''in'' SEP-70, 413--436. Stanford Exploration Project.</ref>). The format is made as simple as possible for maximum convenience, transparency, and flexibility. | ||
maximum convenience, transparency and flexibility. | According to the Unix tradition, standard file formats should be in a readable | ||
According to the Unix tradition, | textual form to be easily examined and processed with universal | ||
textual form | tools. Raymond (2004<ref>Raymond, E. S., 2004, The art of UNIX programming: Addison-Wesley.</ref>) writes: | ||
tools. | |||
<blockquote> | <blockquote> | ||
To design a perfect anti-Unix, make all file formats binary and opaque | To design a perfect anti-Unix, make all file formats binary and opaque and | ||
require heavyweight tools to read and edit them. | require heavyweight tools to read and edit them. | ||
</blockquote> | </blockquote> | ||
<blockquote> | <blockquote> | ||
If you feel an urge to design a complex binary file format | If you feel an urge to design a complex binary file format or a complex | ||
binary application protocol, it is generally wise to lie down until the | binary application protocol, it is generally wise to lie down until the | ||
feeling passes. | feeling passes. | ||
| Line 21: | Line 23: | ||
Storing large-scale datasets in a text format may not be economical. RSF | Storing large-scale datasets in a text format may not be economical. RSF | ||
chooses the next best thing: it allows data values to be stored in a binary | chooses the next best thing: it allows data values to be stored in a binary | ||
format but puts all data attributes in text files that can | format but puts all data attributes in text files that humans can read | ||
and processed with universal text-processing utilities. | and processed with universal text-processing utilities. | ||
===Example=== | ===Example=== | ||
| Line 41: | Line 43: | ||
n1=1000 | n1=1000 | ||
</pre> | </pre> | ||
The file contains nine lines with simple readable text. The first line | The file contains nine lines with simple, readable text. The first line | ||
shows the name of the program, the working directory, the user and | shows the name of the program, the working directory, the user, and | ||
computer that created the file and the time it was created (that | computer that created the file and the time it was created (that | ||
information is recorded for accounting purposes). Other lines contain | information is recorded for accounting purposes). Other lines contain | ||
parameter-value pairs separated by the "=" sign. The "in" | parameter-value pairs separated by the "=" sign. The "in" | ||
parameter points to the location of the binary data. Before we discuss | parameter points to the location of the binary data. Before we discuss the meaning of parameters in more detail, let us plot the data. | ||
the meaning of parameters in more detail, let us plot the data. | |||
<pre> | <pre> | ||
bash$ < sin.rsf sfwiggle title='One Trace' | | bash$ < sin.rsf sfwiggle title='One Trace' | sfpen | ||
</pre> | </pre> | ||
You should see a plot similar to the figure below on your screen. | |||
[[Image:sin1.png|frame|center|An example sinusoid plot.]] | [[Image:sin1.png|frame|center|An example sinusoid plot.]] | ||
Suppose you want to reformat the data so that instead of one trace of a | Suppose you want to reformat the data so that instead of one trace of a thousand samples, it contains twenty traces with fifty samples each. Try | ||
thousand samples, it contains twenty traces with fifty samples each. Try | |||
running | running | ||
<pre> | <pre> | ||
bash$ < sin.rsf sed 's/n1=1000/n1= | bash$ < sin.rsf sed 's/n1=1000/n1=50 n2=20/' > sin10.rsf | ||
bash$ < sin10.rsf sfwiggle title=Traces | | bash$ < sin10.rsf sfwiggle title=Traces | sfpen | ||
</pre> | </pre> | ||
or (using pipes) | or (using pipes) | ||
<pre> | <pre> | ||
bash$ < sin.rsf sed 's/n1=1000/n1=50 n2=20/' | sfwiggle title=Traces | | bash$ < sin.rsf sed 's/n1=1000/n1=50 n2=20/' | sfwiggle title=Traces | sfpen | ||
</pre> | </pre> | ||
On your screen, you should see a plot similar to | On your screen, you should see a plot similar to the figure below: | ||
[[Image:sin2.png|frame|center|An example sinusoid plot, with data reformatted to twenty traces.]] | [[Image:sin2.png|frame|center|An example sinusoid plot, with data reformatted to twenty traces.]] | ||
What happened? We used <tt>sed</tt>, a standard Unix line editing utility to | What happened? We used <tt>sed</tt>, a standard Unix line editing utility, to | ||
change the parameters describing the data dimensions. Because of the | change the parameters describing the data dimensions. Because of the | ||
simplicity of this operation, there is no need to create specialized data | simplicity of this operation, there is no need to create specialized data | ||
| Line 75: | Line 75: | ||
<pre> | <pre> | ||
bash$ ( cat sin.rsf; echo n1=50 n2=20 ) > sin10.rsf | bash$ ( cat sin.rsf; echo n1=50 n2=20 ) > sin10.rsf | ||
bash$ < sin10.rsf sfwiggle title=Traces | | bash$ < sin10.rsf sfwiggle title=Traces | sfpen | ||
</pre> | </pre> | ||
In this case, the <tt>cat</tt> utility | In this case, the <tt>cat</tt> utility copies the contents of the | ||
previous file, and the <tt>echo</tt> utility appends new line "n1=50 | previous file, and the <tt>echo</tt> utility appends a new line "<tt>n1=50 | ||
n2=20". A new value of the <tt>n1</tt> parameter overwrites the old value | n2=20</tt>". A new value of the <tt>n1</tt> parameter overwrites the old value | ||
of <tt>n1=1000</tt>, and we achieve the same result as before. | of <tt>n1=1000</tt>, and we achieve the same result as before. | ||
Of course, one could also edit the file by hand with one of the general | Of course, one could also edit the file by hand with one of the general-purpose text editors. For recording the history of data processing, it is | ||
purpose text editors. For recording the history of data processing, it is | |||
usually preferable to be able to process files with non-interactive tools. | usually preferable to be able to process files with non-interactive tools. | ||
| Line 110: | Line 109: | ||
</pre> | </pre> | ||
4000 bytes in this file are required to store <math>50 \times 20</math> floating-point | 4000 bytes in this file are required to store <math>50 \times 20</math> floating-point | ||
4-byte numbers in a binary form. Thus, the data file contains | 4-byte numbers in a binary form. Thus, the data file contains only the | ||
raw data in a contiguous binary form. | raw data in a contiguous binary form. | ||
===Datapath=== | ===Datapath=== | ||
| Line 117: | Line 116: | ||
data file directory are as follows: | data file directory are as follows: | ||
#Check <tt>out=</tt> parameter on the command line. The parameter specifies the output data file location explicitly. | #Check <tt>--out=</tt> parameter on the command line. The parameter specifies the output data file location explicitly. | ||
#Specify the path and the file name separately. | #Specify the path and the file name separately. | ||
#*The rules for the path selection are: | #*The rules for the path selection are: | ||
#*#Check <tt>datapath=</tt> parameter on the command line. The parameter specifies a string to prepend to the file name. The string may contain the file directory. | #*#Check <tt>datapath=</tt> parameter on the command line. The parameter specifies a string to prepend to the file name. The string may contain the file directory. | ||
#*#Check <tt>DATAPATH</tt> environmental variable. It has the same meaning as the parameter specified with <tt>datapath=</tt>. | #*#Check <tt>DATAPATH</tt> environmental variable. It has the same meaning as the parameter specified with <tt>datapath=</tt>. | ||
#*#Check for <tt>.datapath</tt> file in the current directory. The file may contain a line <pre> datapath=/path/to_file/ </pre> or <pre> machine_name datapath=/path/to_file/ </pre> if you | #*#Check for <tt>.datapath</tt> file in the current directory. The file may contain a line <pre> datapath=/path/to_file/ </pre> or <pre> machine_name datapath=/path/to_file/ </pre> if you intend to use different paths on different platforms. | ||
#*#Check for <tt>.datapath</tt> file in the user home directory. | #*#Check for <tt>.datapath</tt> file in the user's home directory. | ||
#*#Put the data file in the current directory (similar to <tt>datapath=./</tt>). | #*#Put the data file in the current directory (similar to <tt>datapath=./</tt>). | ||
#*: | #*: | ||
#*The rules for the filename selection are: | #*The rules for the filename selection are: | ||
#*#If the output RSF file is in the current directory, the name of the data file is made by appending \@. | #*#If the output RSF file is in the current directory, the name of the data file is made by appending \@. | ||
#*#If the output file is not in the current directory or | #*#If the output file is not in the current directory or is created temporarily by a program, the name is made by appending random characters to the program's name and selected to be unique. | ||
Examples: | Examples: | ||
<pre> | |||
<pre> | bash$ sfspike n1=10 --out=test1 > spike.rsf | ||
bash$ sfspike n1=10 out=test1 > spike.rsf | |||
bash$ grep in spike.rsf | bash$ grep in spike.rsf | ||
in="test1" | in="test1" | ||
</pre> | </pre> | ||
<pre> | <pre> | ||
bash$ sfspike n1=10 datapath=/tmp/ > spike.rsf | bash$ sfspike n1=10 datapath=/tmp/ > spike.rsf | ||
bash$ grep in spike.rsf | bash$ grep in spike.rsf | ||
in="/tmp/spike.rsf@" | in="/tmp/spike.rsf@" | ||
</pre> | </pre> | ||
<pre> | <pre> | ||
bash$ DATAPATH=/tmp/ sfspike n1=10 > spike.rsf | bash$ DATAPATH=/tmp/ sfspike n1=10 > spike.rsf | ||
bash$ grep in spike.rsf | bash$ grep in spike.rsf | ||
in="/tmp/spike.rsf@" | in="/tmp/spike.rsf@" | ||
</pre> | </pre> | ||
<pre> | <pre> | ||
bash$ sfspike n1=10 datapath=/tmp/ > /tmp/spike.rsf | bash$ sfspike n1=10 datapath=/tmp/ > /tmp/spike.rsf | ||
bash$ grep in /tmp/spike.rsf | bash$ grep in /tmp/spike.rsf | ||
| Line 161: | Line 158: | ||
While the header and data files are separated by default, it is also possible | While the header and data files are separated by default, it is also possible | ||
to pack them together into one file. To do that, specify the program's | to pack them together into one file. To do that, specify the program's | ||
"<tt>out</tt>" parameter as <tt>out=stdout</tt>. Example: | "<tt>--out</tt>" parameter as <tt>--out=stdout</tt>. Example: | ||
<pre> | <pre> | ||
bash$ sfspike n1=10 out=stdout > spike.rsf | bash$ sfspike n1=10 --out=stdout > spike.rsf | ||
bash$ grep in spike.rsf | bash$ grep in spike.rsf | ||
Binary file spike.rsf matches | Binary file spike.rsf matches | ||
| Line 178: | Line 175: | ||
starts with the text header information, followed by special | starts with the text header information, followed by special | ||
symbols, followed by binary data. | symbols, followed by binary data. | ||
Packing headers and data together may not be a good idea for data processing | Packing headers and data together may not be a good idea for data processing, but it works well for storing data: it is easier to move the packed file | ||
but it works well for storing data: it is easier to move the packed file | |||
around than to move two different files (header and binary) together while | around than to move two different files (header and binary) together while | ||
remembering to preserve their connection. Packing header and data together is | remembering to preserve their connection. Packing the header and data together is also the current mechanism used to push RSF files through Unix pipes. | ||
also the current mechanism used to push RSF files through Unix pipes. | |||
===Type=== | ===Type=== | ||
| Line 188: | Line 183: | ||
character, integer, floating point, or complex. By default, single precision | character, integer, floating point, or complex. By default, single precision | ||
is used for numbers (<tt>int</tt> and <tt>float</tt> data types in the C | is used for numbers (<tt>int</tt> and <tt>float</tt> data types in the C | ||
programming language). The number of bytes required | programming language), but double precision and other | ||
integer types (<tt>short</tt> and <tt>long</tt>) are also | |||
supported. The number of bytes required to represent these | |||
numbers may depend on the platform. | numbers may depend on the platform. | ||
===Form=== | ===Form=== | ||
The data stored with RSF can also be in | The data stored with RSF can also be in different forms: ASCII, native | ||
binary, and XDR binary. Native binary is often used by default. It is the | binary, and XDR binary. Native binary is often used by default. It is the | ||
binary format employed by the machine | binary format employed by the machine running the application. On | ||
Linux-running PC, the native binary format will typically correspond to the | Linux-running PC, the native binary format will typically correspond to the | ||
so-called little-endian byte ordering. On some other | so-called little-endian byte ordering. On some other platforms, it might be | ||
big-endian ordering. XDR is a binary format designed by Sun for exchanging | big-endian ordering. XDR is a binary format designed by Sun for exchanging | ||
files over network. It typically corresponds to big-endian byte ordering. It | files over the network. It typically corresponds to big-endian byte ordering. It | ||
is more efficient to process RSF files in the native binary format but | is more efficient to process RSF files in the native binary format, but storing the corresponding file in an XDR format might be a good idea if you intend to access data from different platforms. RSF also allows for an ASCII | ||
intend to access data from different platforms | |||
(plain text) form of data files. | (plain text) form of data files. | ||
Conversion between different types and forms is accomplished with | Conversion between different types and forms is accomplished with | ||
<tt>sfdd</tt> program. Here are some examples. First, let us create synthetic | <tt>sfdd</tt> program. Here are some examples. First, let us create synthetic data. | ||
data. | |||
<pre> | <pre> | ||
bash$ sfmath n1=10 output='10*sin(0.5*x1)' > sin.rsf | bash$ sfmath n1=10 output='10*sin(0.5*x1)' > sin.rsf | ||
| Line 250: | Line 244: | ||
data model is a multidimensional hypercube. By convention, the | data model is a multidimensional hypercube. By convention, the | ||
dimensions of the cube are defined with parameters <tt>n1</tt>, | dimensions of the cube are defined with parameters <tt>n1</tt>, | ||
<tt>n2</tt>, <tt>n3</tt>, etc. | <tt>n2</tt>, <tt>n3</tt>, etc. The fastest axis is <tt>n1</tt>. | ||
Additionally, the grid sampling can be given by parameters | Additionally, the grid sampling can be given by parameters | ||
<tt>d1</tt>, <tt>d2</tt>, <tt>d3</tt>, etc. The axes origins are given | <tt>d1</tt>, <tt>d2</tt>, <tt>d3</tt>, etc. The axes origins are given | ||
by parameters <tt>o1</tt>, <tt>o2</tt>, <tt>o3</tt>, etc. Optionally, | by parameters <tt>o1</tt>, <tt>o2</tt>, <tt>o3</tt>, etc. Optionally, | ||
you can also supply the axis label strings <tt>label1</tt>, | you can also supply the axis label strings: <tt>label1</tt>, | ||
<tt>label2</tt>, <tt>label3</tt>, etc., and axis units strings | <tt>label2</tt>, <tt>label3</tt>, etc., and axis units strings: | ||
<tt>unit1</tt>, <tt>unit2</tt>, <tt>unit3</tt>, etc. | <tt>unit1</tt>, <tt>unit2</tt>, <tt>unit3</tt>, etc. | ||
==Compatibility with other file formats== | ==Compatibility with other file formats== | ||
It is possible to exchange RSF-formatted data with other popular data formats. | It is possible to exchange RSF-formatted data with several other popular data formats. | ||
===Compatibility with SEPlib=== | ===Compatibility with SEPlib=== | ||
RSF is mostly compatible with its predecessor, the SEPlib file format. | RSF is mostly compatible with its predecessor, the SEPlib file format. | ||
However, there are several | However, there are several significant differences: | ||
#SEPlib | #SEPlib programs typically use the element size (<tt>esize=</tt> parameter) to distinguish between different data types: <tt>esize=4</tt> corresponds to floating point data, while <tt>esize=8</tt> corresponds to complex data. The RSF type handling mechanism is different: data types are determined from the value of the <tt>data_format</tt> parameter. Madagascar computational programs typically output files with <tt>data_format="native_float"</tt> or <tt>native_complex</tt>. | ||
#The default data form in SEPlib programs is typically XDR and not native as it is in RSF. | #The default data form in SEPlib programs is typically XDR and not native as it is in RSF. Thus, to make a dataset created with SEPlib readable by Madagascar programs, you would typically need to add to the history file <tt>data_format="xdr_float"</tt> or <tt>data_format="xdr_complex"</tt> . <ref group="note">For SEPlib 6.5.3 and older: Note that xdr_complex is not a valid SEPlib value, so for datasets of complex numbers encoded as pairs of floats, a dataset cannot be at the same time valid in both SEPlib and Madagascar. A valid SEPlib dataset will have esize=8 and data_format="xdr_float", but sfin will show it as having "200% of expected" data. Adding data_format="xdr_complex" to such a dataset will make sfin work as expected, but SEPlib's In or In3d will give a segmentation fault because of an unknown data type. To patch SEPlib to accept <tt>native_complex</tt> and <tt>xdr_complex</tt> data, the following changes must be made: | ||
#It is possible to pipe the output of | * In <tt>$SEPSRC/seplib_base/lib/corelibs/sep/strformats.c</tt>: | ||
#SEP3D is an extension of SEPlib for operating with irregularly sampled data (Biondi et al., 1996<ref>Biondi, B., R. Clapp, and S. Crawley, 1996, SEPlib90: SEPlib for 3-D prestack data, ''in'' SEP-92, 343--364. Stanford Exploration Project.</ref>). There is no equivalent of it in RSF for the reasons explained | ** Add "xdr_complex" and "native_complex" to the str_fmt_names structure | ||
** Set FMT_LENGTH to 15 | |||
* In <tt>$SEPSRC/seplib_base/lib/corelibs/include/strformats.h</tt>: | |||
** Add preprocessor directives to define FMT_XDR_COMPLEX as 8 and FMT_NATIVE_COMPLEX as 9 | |||
** Set NUM_FMT to 10 | |||
</ref> | |||
#It is possible to pipe the output of Madagascar programs to SEPlib: <pre>bash$ sfspike n1=1 | Attr want=min</pre> (output should be: <tt>minimum value = 1 at 1</tt>). However, piping the output of SEPlib programs to RSF (or, for that matter, any other non-SEPlib programs) will result in an unterminated process. For example, the command <pre> bash$ Spike n1=1 | sfattr want=min </pre> will hang. This is because SEPlib uses sockets for piping and expects a socket connection from the receiving program, while Madagascar passes data through regular Unix pipes. | |||
#SEP3D is an extension of SEPlib for operating with irregularly sampled data (Biondi et al., 1996<ref>Biondi, B., R. Clapp, and S. Crawley, 1996, SEPlib90: SEPlib for 3-D prestack data, ''in'' SEP-92, 343--364. Stanford Exploration Project.</ref>). There is no equivalent of it in RSF for the reasons explained at the beginning of this guide. Operations with irregular datasets are supported using auxiliary input files representing the geometry information. | |||
;Notes | |||
<references group="note" /> | |||
===Reading and writing SEG-Y and SU files=== | ===Reading and writing SEG-Y and SU files=== | ||
The SEG-Y format is based on the proposal of Barry et al. (1975<ref>Barry, K. M., D. A. Cavers, and C. W. Kneale, 1975, Report on recommended | The SEG-Y format is based on the proposal of Barry et al. (1975<ref>[http://www.seg.org/SEGportalWEBproject/prod/SEG-Publications/Pub-Technical-Standards/Documents/seg_y_rev0.pdf Barry, K. M., D. A. Cavers, and C. W. Kneale, 1975, Report on recommended standards for digital tape formats: Geophysics, '''40''', 344--352]</ref>). | ||
It was revised in 2002<ref>See http://seg.org/ | It was revised in 2002<ref>See http://www.seg.org/SEGportalWEBproject/prod/SEG-Publications/Pub-Technical-Standards/Documents/seg_y_rev1.pdf</ref>. The | ||
SU format is a modification of SEG-Y used in Seismic Unix | SU format is a modification of SEG-Y used in Seismic Unix | ||
(Stockwell, 1997<ref>Stockwell, J. W., 1997, Free software in education: A case study of CWP/SU: Seismic Unix: The Leading Edge, '''16''', 1045--1049.</ref>). | (Stockwell, 1997<ref>Stockwell, J. W., 1997, Free software in education: A case study of CWP/SU: Seismic Unix: The Leading Edge, '''16''', 1045--1049.</ref>). | ||
| Line 286: | Line 291: | ||
<pre> | <pre> | ||
bash$ sfsegyread < plane.segy tfile=tfile.rsf \ | bash$ sfsegyread < plane.segy tfile=tfile.rsf \ | ||
hfile= | hfile=file.asc bfile=file.bin > plane.rsf | ||
</pre> | </pre> | ||
The endian flag is needed if the SU file originated from a little-endian | The endian flag is needed if the SU file originated from a little-endian machine like a Linux PC. | ||
machine | |||
Several files are generated. The standard output contains an RSF file with the | Several files are generated. The standard output contains an RSF file with the | ||
data (32 traces with 64 samples each): | data (32 traces with 64 samples each): | ||
| Line 317: | Line 321: | ||
The contents of trace headers can be quickly examined with the | The contents of trace headers can be quickly examined with the | ||
<tt>sfheaderattr</tt> program. | <tt>sfheaderattr</tt> program. | ||
The <tt> | The <tt>file.asc</tt> is the ASCII header file for the whole record. | ||
<pre> | <pre> | ||
bash$ head -c 242 | bash$ head -c 242 file.asc | ||
C This tape was made at the | C This tape was made at the | ||
C | C | ||
C Center for Wave Phenomena | C Center for Wave Phenomena | ||
</pre> | </pre> | ||
The <tt> | The <tt>file.bin</tt> is the binary header file. | ||
To convert files back from RSF to SEG-Y or SU, use the <tt>sfsegywrite</tt> | To convert files back from RSF to SEG-Y or SU, use the <tt>sfsegywrite</tt> | ||
program and reverse the input and output: | program and reverse the input and output: | ||
<pre> | <pre> | ||
bash$ sfsuwrite > | bash$ sfsuwrite > plane.su tfile=tfile.rsf endian=0 < plane.rsf | ||
</pre> | </pre> | ||
or | or | ||
<pre> | <pre> | ||
bash$ sfsegywrite > | bash$ sfsegywrite > plane.segy tfile=tfile.rsf \ | ||
hfile= | hfile=file.asc bfile=file.bin < plane.rsf | ||
</pre> | </pre> | ||
If <tt>hfile=</tt> and <tt>bfile=</tt> are not supplied to <tt>sfsegywrite</tt>, the corresponding headers will be | If <tt>hfile=</tt> and <tt>bfile=</tt> are not supplied to <tt>sfsegywrite</tt>, the corresponding headers will be generated on the fly. The trace header file can be generated with <tt>sfsegyheader</tt>. Here is an example: | ||
<pre> | <pre> | ||
bash$ sfheadermath < plane.rsf output=N+1 | sfdd type=int > tracl.rsf | |||
bash$ sfheadermath < | bash$ sfsegyheader < plane.rsf tracl=tracl.rsf > tfile.rsf | ||
bash$ sfsegyheader < | bash$ sfsegywrite < plane.rsf tfile=tfile.rsf > plane.segy | ||
bash$ sfsegywrite < | |||
</pre> | </pre> | ||
====Unusual trace header keys==== | |||
Sometimes, SEG-Y files deviate from the standard by creating additional | |||
trace header keys. If, for example, you find out that the SEG-Y file | |||
contains an additional trace header key stored in bytes 225-226, you can either remap one of the standard two-byte keys | |||
<pre> | |||
bash$ sfsegyread < file.segy tfile=tfile.rsf gut=224 > file.rsf | |||
</pre> | |||
or create a new key | |||
<pre> | |||
bash$ sfsegyread < file.segy tfile=tfile.rsf \ | |||
key1=mykey key1_len=2 mykey=224 > file.rsf | |||
</pre> | |||
Any number of additional keys can be created this way. | |||
===Reading and writing ASCII files=== | ===Reading and writing ASCII files=== | ||
| Line 377: | Line 394: | ||
6 elements 24 bytes | 6 elements 24 bytes | ||
</pre> | </pre> | ||
Converting from RSF to ASCII is equally simple: | |||
<pre> | <pre> | ||
bash$ sfdd form=ascii out=file.asc < file.rsf > /dev/null | bash$ sfdd form=ascii --out=file.asc < file.rsf > /dev/null | ||
bash$ cat file.asc | bash$ cat file.asc | ||
1 1.5 3 4.8 9.1 7.3 | 1 1.5 3 4.8 9.1 7.3 | ||
| Line 386: | Line 403: | ||
<tt>sfdd</tt> to control the ASCII formatting: | <tt>sfdd</tt> to control the ASCII formatting: | ||
<pre> | <pre> | ||
bash$ sfdd form=ascii out=file.asc \ | bash$ sfdd form=ascii --out=file.asc \ | ||
line=3 format="%3.1f " < file.rsf > /dev/null | line=3 format="%3.1f " < file.rsf > /dev/null | ||
bash$ cat file.asc | bash$ cat file.asc | ||
| Line 399: | Line 416: | ||
4.8 9.1 7.3 | 4.8 9.1 7.3 | ||
</pre> | </pre> | ||
===Reading and writing CSV files=== | |||
CSV (Comma-separated values) is a particular example of an ASCII | |||
format, where commas separate values on different rows or | |||
other symbols. To convert from CSV to RSF, you can use the | |||
<tt>sfcsv2rsf</tt> utility. | |||
For example, let us take an ASCII file with numbers separated by commas | |||
<pre> | |||
bash$ cat file.csv | |||
1.0,1.5,3.0 | |||
4.8,9.1,7.3 | |||
</pre> | |||
Converting it to RSF: | |||
<pre> | |||
bash$ sfcsv2rsf < file.csv > file.rsf | |||
bash$ sfin file.rsf | |||
file.rsf: | |||
in="/tmp/file.rsf@" | |||
esize=4 type=float form=native | |||
n1=3 d1=1 o1=0 label1="unknown" unit1="unknown" | |||
n2=2 d2=1 o2=0 label2="unknown" unit2="unknown" | |||
6 elements 24 bytes | |||
</pre> | |||
To convert from RSF to CSV, we can use formatting parameters in <tt>sfdd</tt>: | |||
<pre> | |||
bash$ sfdd form=ascii --out=file.csv \ | |||
line=3 strip=1 format="%3.1f," < file.rsf >/dev/null | |||
bash$ cat file.csv | |||
1.0,1.5,3.0 | |||
4.8,9.1,7.3 | |||
</pre> | |||
Some CSV files contain headers with definitions for different columns. | |||
<pre> | |||
bash$ cat file.csv | |||
height,width,weight | |||
1.0,1.5,3.0 | |||
4.8,9.1,7.3 | |||
</pre> | |||
To read a file like that, use <tt>header=</tt> parameter in <tt>sfcsv2rsf</tt>, as follows: | |||
<pre> | |||
bash$ sfcsv2rsf < file.csv header=y > file.rsf | |||
</pre> | |||
After that, different columns can be accessed by keywords. | |||
<pre> | |||
bash$ < file.rsf sfheaderattr segy=n | |||
3 headers, 2 traces | |||
******************************************************************************* | |||
key min max mean | |||
------------------------------------------------------------------------------- | |||
height 0 1 @ 0 4.8 @ 1 2.9 | |||
width 1 1.5 @ 0 9.1 @ 1 5.3 | |||
weight 2 3 @ 0 7.3 @ 1 5.15 | |||
******************************************************************************* | |||
</pre> | |||
===Reading LAS files=== | |||
LAS (Log ASCII Standard) is a text format used for storing | |||
well-logging data (Heslop et al., 1999<ref>Heslop, K., J. Karst, S. Prensky, D. Schmitt, et al., 1999, Log ASCII standard LAS version 3.0: The Log Analyst, 40.</ref>). LAS files can be converted to the RSF format using | |||
<tt>sflas2rsf</tt> utility. | |||
Let us try an example file from one of the SEG tutorials: | |||
<pre> | |||
bash$ tutorials=https://raw.githubusercontent.com/seg/tutorials-2014/master | |||
bash$ wget $tutorials/1406_Make_a_synthetic/L-30.las | |||
</pre> | |||
Converting to RSF, we can detect 15 different logs: | |||
<pre> | |||
bash$ sflas2rsf L-30.las L-30.rsf | |||
(base) sergey@DESKTOP-80QRDA0:~/all/fomels/nnint$ sfin L-30.rsf | |||
L-30.rsf: | |||
in="/home/sergey/RSFROOT/data/L-30.rsf@" | |||
esize=4 type=float form=native | |||
n1=15 d1=? o1=? | |||
n2=25621 d2=0.5 o2=1140 | |||
384315 elements 1537260 bytes | |||
</pre> | |||
Individual logs are accessible by their keys and can be used in programs like <tt>sfheadermath</tt>. | |||
<pre> | |||
bash$ < L-30.rsf sfheaderattr segy=n desc=y | |||
15 headers, 25621 traces | |||
******************************************************************************* | |||
key min max mean | |||
------------------------------------------------------------------------------- | |||
DEPTH 0 1140 @ 0 13950 @ 25620 7545 | |||
[Depth] | |||
CALD 1 -999 @ 0 19.811 @ 3909 -140.356 | |||
[Caliper Caliper - Density] | |||
CALS 2 -999 @ 0 14.84 @ 23096 7.43849 | |||
[Caliper Caliper - Sonic] | |||
DEPT 3 1140 @ 0 13950 @ 25620 7545 | |||
[Depth] | |||
DRHO 4 -999 @ 0 0.254 @ 23667 -149.67 | |||
[Drho Delta Rho] | |||
DT 5 -999 @ 0 199.263 @ 1462 90.0167 | |||
[Sonic Delta-T] | |||
GRD 6 -999 @ 0 178.416 @ 21788 -100.952 | |||
[GammaRay Gamma Ray - Density] | |||
GRS 7 -999 @ 0 140.148 @ 23376 53.8002 | |||
[GammaRay Gamma Ray - Sonic] | |||
ILD 8 -999 @ 0 2022.95 @ 20 34.5917 | |||
[DeepRes Deep Induction Standard Processed Resistivity] | |||
ILM 9 -999 @ 0 2196.26 @ 20661 40.5595 | |||
[MedRes Medium Induction Standard Processed Resistivity] | |||
LL8 10 -999 @ 0 2097.76 @ 20213 35.6343 | |||
[ShalRes Latero-Log 8] | |||
NPHILS 11 -999 @ 0 0.45 @ 23039 -776.522 | |||
[Neutron Neutron Porosity - Ls Mtx] | |||
NPHISS 12 -999 @ 0 0.615 @ 5215 -373.244 | |||
[Neutron Neutron Porosity - Ss Mtx] | |||
RHOB 13 -999 @ 0 2.811 @ 23941 -147.773 | |||
[Density Bulk Density] | |||
SP 14 -999 @ 0 -19.065 @ 20570 -105.029 | |||
[SP Spontaneous Potential] | |||
******************************************************************************* | |||
bash$ < L-30.rsf sfheadermath output=RHOB segy=n > RHOB.rsf | |||
bash$ < RHOB.rsf sfwindow min2=4000 max2=13000 | sfgraph title=Density | |||
</pre> | |||
[[Image:rhob.png|frame|center|Density log.]] | |||
==Other documentation== | ==Other documentation== | ||
This note should give you a general understanding of the RSF file | This note should give you a general understanding of the RSF file format. See the [[RSF Comprehensive Description]] if you want minutia. Other relevant documentation is: | ||
format. Other relevant documentation is | |||
*[[Why Madagascar]] | *[[Why Madagascar]] | ||
*[[Installation|Installation instructions]] | *[[Installation|Installation instructions]] | ||
*[ | *[https://ahay.org/RSF/ Madagascar self-documentation] | ||
*[[Guide to madagascar programs]] | *[[Guide to madagascar programs]] | ||
*[[Guide to madagascar API|Guide to the Madagascar programming interface]] | *[[Guide to madagascar API|Guide to the Madagascar programming interface]] | ||
| Line 413: | Line 546: | ||
*[[Revisiting SEP tour with Madagascar and SCons]] | *[[Revisiting SEP tour with Madagascar and SCons]] | ||
*[[Reproducible computational experiments using SCons]] | *[[Reproducible computational experiments using SCons]] | ||
==About this document== | |||
This page was created from the LaTeX source in [http://rsf.svn.sourceforge.net/viewvc/rsf/trunk/book/rsf/rsf/format.tex?view=markup book/rsf/rsf/format.tex] using [[latex2wiki]]. | |||
==References== | ==References== | ||
<references/> | <references/> | ||
Latest revision as of 19:58, 20 November 2024

Principles[edit]
The main design principle behind the RSF data format is KISS ("Keep It Short and Simple"). The RSF format is borrowed from the SEPlib data format initially designed at the Stanford Exploration Project (Claerbout, 1991[1]). The format is made as simple as possible for maximum convenience, transparency, and flexibility. According to the Unix tradition, standard file formats should be in a readable textual form to be easily examined and processed with universal tools. Raymond (2004[2]) writes:
To design a perfect anti-Unix, make all file formats binary and opaque and require heavyweight tools to read and edit them.
If you feel an urge to design a complex binary file format or a complex binary application protocol, it is generally wise to lie down until the feeling passes.
Storing large-scale datasets in a text format may not be economical. RSF chooses the next best thing: it allows data values to be stored in a binary format but puts all data attributes in text files that humans can read and processed with universal text-processing utilities.
Example[edit]
Let us first create some synthetic RSF data.
bash$ sfmath n1=1000 output='sin(0.5*x1)' > sin.rsf
Open and read the file sin.rsf.
bash$ cat sin.rsf
sfmath rsf/rsf/rsftour: fomels@egl Sun Jul 31 07:18:48 2005
o1=0
data_format="native_float"
esize=4
in="/tmp/sin.rsf@"
x1=0
d1=1
n1=1000
The file contains nine lines with simple, readable text. The first line shows the name of the program, the working directory, the user, and computer that created the file and the time it was created (that information is recorded for accounting purposes). Other lines contain parameter-value pairs separated by the "=" sign. The "in" parameter points to the location of the binary data. Before we discuss the meaning of parameters in more detail, let us plot the data.
bash$ < sin.rsf sfwiggle title='One Trace' | sfpen
You should see a plot similar to the figure below on your screen.
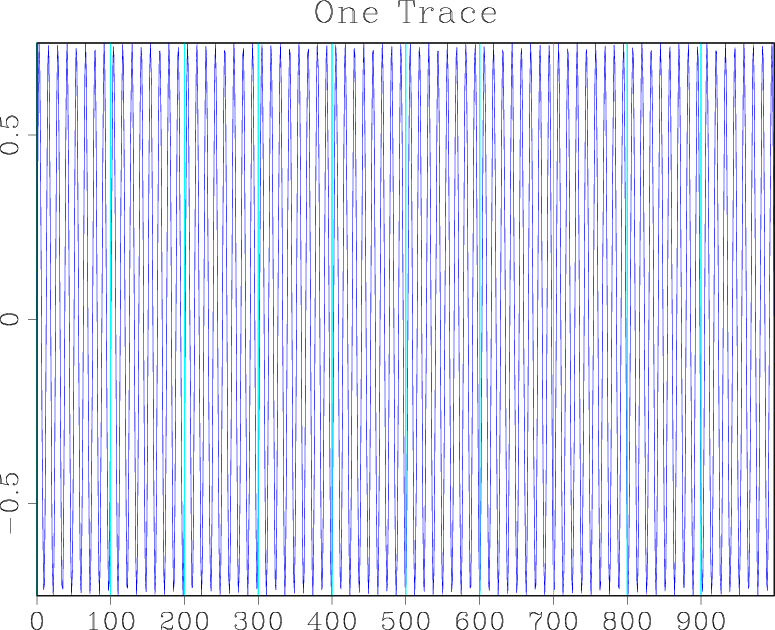
Suppose you want to reformat the data so that instead of one trace of a thousand samples, it contains twenty traces with fifty samples each. Try running
bash$ < sin.rsf sed 's/n1=1000/n1=50 n2=20/' > sin10.rsf bash$ < sin10.rsf sfwiggle title=Traces | sfpen
or (using pipes)
bash$ < sin.rsf sed 's/n1=1000/n1=50 n2=20/' | sfwiggle title=Traces | sfpen
On your screen, you should see a plot similar to the figure below:
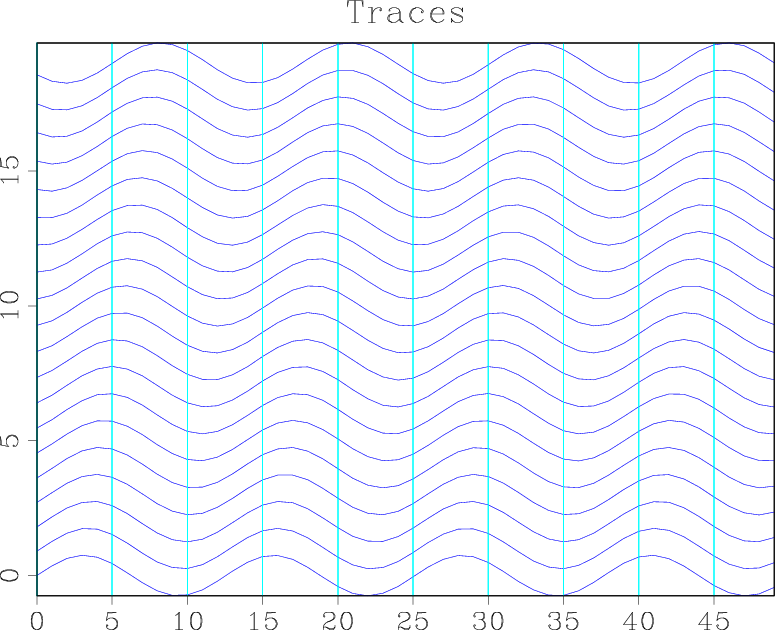
What happened? We used sed, a standard Unix line editing utility, to change the parameters describing the data dimensions. Because of the simplicity of this operation, there is no need to create specialized data formatting tools or to make the sfwiggle program accept additional formatting parameters. Other general-purpose Unix tools that can be applied on RSF files include cat, echo, grep, etc. An alternative way to obtain the previous result is to run
bash$ ( cat sin.rsf; echo n1=50 n2=20 ) > sin10.rsf bash$ < sin10.rsf sfwiggle title=Traces | sfpen
In this case, the cat utility copies the contents of the previous file, and the echo utility appends a new line "n1=50 n2=20". A new value of the n1 parameter overwrites the old value of n1=1000, and we achieve the same result as before. Of course, one could also edit the file by hand with one of the general-purpose text editors. For recording the history of data processing, it is usually preferable to be able to process files with non-interactive tools.
Header and Data files[edit]
A simple way to check the layout of an RSF file is with the sfin program.
bash$ sfin sin10.rsf
sin10.rsf:
in="/tmp/sin.rsf@"
esize=4 type=float form=native
n1=50 d1=1 o1=0
n2=20 d2=? o2=?
1000 elements 4000 bytes
The program reports the following information: the location of the data file (/tmp/sin.rsf\@), the element size (4 bytes), the element type (floating point), the element form (native), the hypercube dimensions (), axis scaling (1 and unspecified), and axis origin (0 and unspecified). It also checks the total number of elements and bytes in the data file. Let us examine this information in detail. First, we can verify that the data file exists and contains the specified number of bytes:
bash$ ls -l /tmp/sin.rsf@ -rw-r--r-- 1 sergey users 4000 2004-10-04 00:35 /tmp/sin.rsf@
4000 bytes in this file are required to store floating-point 4-byte numbers in a binary form. Thus, the data file contains only the raw data in a contiguous binary form.
Datapath[edit]
How did the RSF program (sfmath) decide where to put the data file? In the order of priority, the rules for selecting the data file name and the data file directory are as follows:
- Check --out= parameter on the command line. The parameter specifies the output data file location explicitly.
- Specify the path and the file name separately.
- The rules for the path selection are:
- Check datapath= parameter on the command line. The parameter specifies a string to prepend to the file name. The string may contain the file directory.
- Check DATAPATH environmental variable. It has the same meaning as the parameter specified with datapath=.
- Check for .datapath file in the current directory. The file may contain a line
datapath=/path/to_file/
ormachine_name datapath=/path/to_file/
if you intend to use different paths on different platforms. - Check for .datapath file in the user's home directory.
- Put the data file in the current directory (similar to datapath=./).
- The rules for the filename selection are:
- If the output RSF file is in the current directory, the name of the data file is made by appending \@.
- If the output file is not in the current directory or is created temporarily by a program, the name is made by appending random characters to the program's name and selected to be unique.
- The rules for the path selection are:
Examples:
bash$ sfspike n1=10 --out=test1 > spike.rsf bash$ grep in spike.rsf in="test1"
bash$ sfspike n1=10 datapath=/tmp/ > spike.rsf bash$ grep in spike.rsf in="/tmp/spike.rsf@"
bash$ DATAPATH=/tmp/ sfspike n1=10 > spike.rsf bash$ grep in spike.rsf in="/tmp/spike.rsf@"
bash$ sfspike n1=10 datapath=/tmp/ > /tmp/spike.rsf bash$ grep in /tmp/spike.rsf in="/tmp/sfspikejcARVf"
Packing header and data together[edit]
While the header and data files are separated by default, it is also possible to pack them together into one file. To do that, specify the program's "--out" parameter as --out=stdout. Example:
bash$ sfspike n1=10 --out=stdout > spike.rsf
bash$ grep in spike.rsf
Binary file spike.rsf matches
bash$ sfin spike.rsf
spike.rsf:
in="stdin"
esize=4 type=float form=native
n1=10 d1=0.004 o1=0 label1="Time" unit1="s"
10 elements 40 bytes
bash$ ls -l spike.rsf
-rw-r--r-- 1 sergey users 196 2004-11-10 21:39 spike.rsf
If you examine the contents of spike.rsf, you will find that it starts with the text header information, followed by special symbols, followed by binary data. Packing headers and data together may not be a good idea for data processing, but it works well for storing data: it is easier to move the packed file around than to move two different files (header and binary) together while remembering to preserve their connection. Packing the header and data together is also the current mechanism used to push RSF files through Unix pipes.
Type[edit]
The data stored with RSF can have different types: character, unsigned character, integer, floating point, or complex. By default, single precision is used for numbers (int and float data types in the C programming language), but double precision and other integer types (short and long) are also supported. The number of bytes required to represent these numbers may depend on the platform.
Form[edit]
The data stored with RSF can also be in different forms: ASCII, native binary, and XDR binary. Native binary is often used by default. It is the binary format employed by the machine running the application. On Linux-running PC, the native binary format will typically correspond to the so-called little-endian byte ordering. On some other platforms, it might be big-endian ordering. XDR is a binary format designed by Sun for exchanging files over the network. It typically corresponds to big-endian byte ordering. It is more efficient to process RSF files in the native binary format, but storing the corresponding file in an XDR format might be a good idea if you intend to access data from different platforms. RSF also allows for an ASCII (plain text) form of data files. Conversion between different types and forms is accomplished with sfdd program. Here are some examples. First, let us create synthetic data.
bash$ sfmath n1=10 output='10*sin(0.5*x1)' > sin.rsf
bash$ sfin sin.rsf
sin.rsf:
in="/tmp/sin.rsf@"
esize=4 type=float form=native
n1=10 d1=1 o1=0
10 elements 40 bytes
bash$ < sin.rsf sfdisfil
0: 0 4.794 8.415 9.975 9.093
5: 5.985 1.411 -3.508 -7.568 -9.775
Converting the data to the integer type:
bash$ < sin.rsf sfdd type=int > isin.rsf
bash$ sfin isin.rsf
isin.rsf:
in="/tmp/isin.rsf@"
esize=4 type=int form=native
n1=10 d1=1 o1=0
10 elements 40 bytes
bash$ < isin.rsf sfdisfil
0: 0 4 8 9 9 5 1 -3 -7 -9
Converting the data to the ASCII form:
bash$ < sin.rsf sfdd form=ascii > asin.rsf
bash$ < asin.rsf sfdisfil
0: 0 4.794 8.415 9.975 9.093
5: 5.985 1.411 -3.508 -7.568 -9.775
bash$ sfin asin.rsf
asin.rsf:
in="/tmp/asin.rsf@"
esize=0 type=float form=ascii
n1=10 d1=1 o1=0
10 elements
bash$ cat /tmp/asin.rsf@
0 4.79426 8.41471 9.97495 9.09297 5.98472 1.4112 -3.50783
-7.56803 -9.7753
Hypercube[edit]
While RSF stores binary data in a contiguous 1-D array, the conceptual data model is a multidimensional hypercube. By convention, the dimensions of the cube are defined with parameters n1, n2, n3, etc. The fastest axis is n1. Additionally, the grid sampling can be given by parameters d1, d2, d3, etc. The axes origins are given by parameters o1, o2, o3, etc. Optionally, you can also supply the axis label strings: label1, label2, label3, etc., and axis units strings: unit1, unit2, unit3, etc.
Compatibility with other file formats[edit]
It is possible to exchange RSF-formatted data with several other popular data formats.
Compatibility with SEPlib[edit]
RSF is mostly compatible with its predecessor, the SEPlib file format. However, there are several significant differences:
- SEPlib programs typically use the element size (esize= parameter) to distinguish between different data types: esize=4 corresponds to floating point data, while esize=8 corresponds to complex data. The RSF type handling mechanism is different: data types are determined from the value of the data_format parameter. Madagascar computational programs typically output files with data_format="native_float" or native_complex.
- The default data form in SEPlib programs is typically XDR and not native as it is in RSF. Thus, to make a dataset created with SEPlib readable by Madagascar programs, you would typically need to add to the history file data_format="xdr_float" or data_format="xdr_complex" . [note 1]
- It is possible to pipe the output of Madagascar programs to SEPlib:
bash$ sfspike n1=1 | Attr want=min
(output should be: minimum value = 1 at 1). However, piping the output of SEPlib programs to RSF (or, for that matter, any other non-SEPlib programs) will result in an unterminated process. For example, the commandbash$ Spike n1=1 | sfattr want=min
will hang. This is because SEPlib uses sockets for piping and expects a socket connection from the receiving program, while Madagascar passes data through regular Unix pipes. - SEP3D is an extension of SEPlib for operating with irregularly sampled data (Biondi et al., 1996[3]). There is no equivalent of it in RSF for the reasons explained at the beginning of this guide. Operations with irregular datasets are supported using auxiliary input files representing the geometry information.
- Notes
- ↑ For SEPlib 6.5.3 and older: Note that xdr_complex is not a valid SEPlib value, so for datasets of complex numbers encoded as pairs of floats, a dataset cannot be at the same time valid in both SEPlib and Madagascar. A valid SEPlib dataset will have esize=8 and data_format="xdr_float", but sfin will show it as having "200% of expected" data. Adding data_format="xdr_complex" to such a dataset will make sfin work as expected, but SEPlib's In or In3d will give a segmentation fault because of an unknown data type. To patch SEPlib to accept native_complex and xdr_complex data, the following changes must be made:
- In $SEPSRC/seplib_base/lib/corelibs/sep/strformats.c:
- Add "xdr_complex" and "native_complex" to the str_fmt_names structure
- Set FMT_LENGTH to 15
- In $SEPSRC/seplib_base/lib/corelibs/include/strformats.h:
- Add preprocessor directives to define FMT_XDR_COMPLEX as 8 and FMT_NATIVE_COMPLEX as 9
- Set NUM_FMT to 10
- In $SEPSRC/seplib_base/lib/corelibs/sep/strformats.c:
Reading and writing SEG-Y and SU files[edit]
The SEG-Y format is based on the proposal of Barry et al. (1975[4]). It was revised in 2002[5]. The SU format is a modification of SEG-Y used in Seismic Unix (Stockwell, 1997[6]). To convert files from SEG-Y or SU format to RSF, use the sfsegyread program. Let us first manufacture an example file using SU utilities (Stockwell, 1999[7]):
bash$ suplane > plane.su bash$ segyhdrs < plane.su | segywrite tape=plane.segy
To convert it to RSF, use either
bash$ sfsuread < plane.su tfile=tfile.rsf endian=0 > plane.rsf
or
bash$ sfsegyread < plane.segy tfile=tfile.rsf \ hfile=file.asc bfile=file.bin > plane.rsf
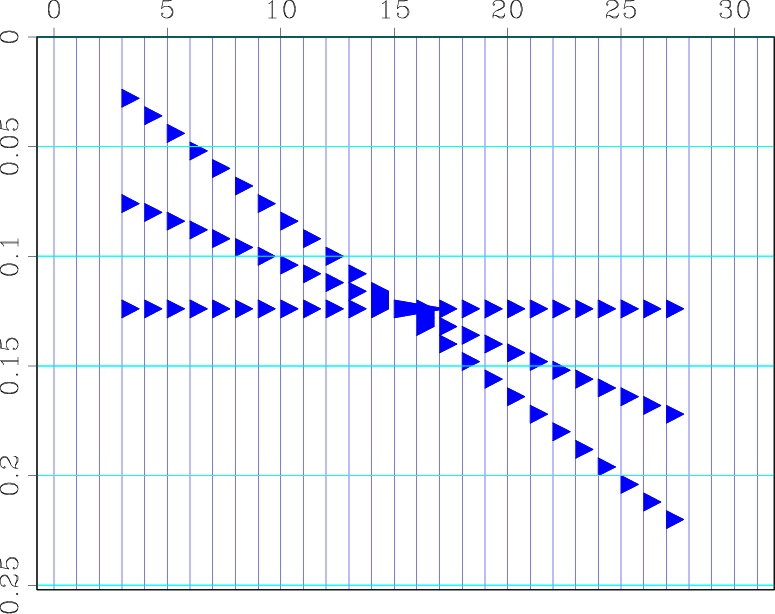
The endian flag is needed if the SU file originated from a little-endian machine like a Linux PC. Several files are generated. The standard output contains an RSF file with the data (32 traces with 64 samples each):
bash$ sfin plane.rsf
plane.rsf:
in="/tmp/plane.rsf@"
esize=4 type=float form=native
n1=64 d1=0.004 o1=0
n2=32 d2=? o2=?
2048 elements 8192 bytes
The contents of this file are displayed in the figure.
The tfile is an RSF integer-type file with the trace headers (32 headers with 71 traces each):
bash$ sfin tfile.rsf
tfile.rsf:
in="/tmp/tfile.rsf@"
esize=4 type=int form=native
n1=71 d1=? o1=?
n2=32 d2=? o2=?
2272 elements 9088 bytes
The contents of trace headers can be quickly examined with the sfheaderattr program. The file.asc is the ASCII header file for the whole record.
bash$ head -c 242 file.asc C This tape was made at the C C Center for Wave Phenomena
The file.bin is the binary header file.
To convert files back from RSF to SEG-Y or SU, use the sfsegywrite program and reverse the input and output:
bash$ sfsuwrite > plane.su tfile=tfile.rsf endian=0 < plane.rsf
or
bash$ sfsegywrite > plane.segy tfile=tfile.rsf \ hfile=file.asc bfile=file.bin < plane.rsf
If hfile= and bfile= are not supplied to sfsegywrite, the corresponding headers will be generated on the fly. The trace header file can be generated with sfsegyheader. Here is an example:
bash$ sfheadermath < plane.rsf output=N+1 | sfdd type=int > tracl.rsf bash$ sfsegyheader < plane.rsf tracl=tracl.rsf > tfile.rsf bash$ sfsegywrite < plane.rsf tfile=tfile.rsf > plane.segy
Unusual trace header keys[edit]
Sometimes, SEG-Y files deviate from the standard by creating additional trace header keys. If, for example, you find out that the SEG-Y file contains an additional trace header key stored in bytes 225-226, you can either remap one of the standard two-byte keys
bash$ sfsegyread < file.segy tfile=tfile.rsf gut=224 > file.rsf
or create a new key
bash$ sfsegyread < file.segy tfile=tfile.rsf \ key1=mykey key1_len=2 mykey=224 > file.rsf
Any number of additional keys can be created this way.
Reading and writing ASCII files[edit]
Reading and writing ASCII files can be accomplished with the sfdd program. For example, let us take an ASCII file with numbers
bash$ cat file.asc 1.0 1.5 3.0 4.8 9.1 7.3
Converting it to RSF is as simple as
bash$ echo in=file.asc n1=3 n2=2 data_format=ascii_float > file.rsf
bash$ sfin file.rsf
file.rsf:
in="file.asc"
esize=0 type=float form=ascii
n1=3 d1=? o1=?
n2=2 d2=? o2=?
6 elements
For more efficient input/output operations, it might be advantageous to convert the data type to native binary, as follows:
bash$ echo in=file.asc n1=3 n2=2 data_format=ascii_float | \
sfdd form=native > file.rsf
bash$ sfin file.rsf
file.rsf:
in="/tmp/file.rsf@"
esize=4 type=float form=native
n1=3 d1=? o1=?
n2=2 d2=? o2=?
6 elements 24 bytes
Converting from RSF to ASCII is equally simple:
bash$ sfdd form=ascii --out=file.asc < file.rsf > /dev/null bash$ cat file.asc 1 1.5 3 4.8 9.1 7.3
You can use the line= and format= parameters in sfdd to control the ASCII formatting:
bash$ sfdd form=ascii --out=file.asc \ line=3 format="%3.1f " < file.rsf > /dev/null bash$ cat file.asc 1.0 1.5 3.0 4.8 9.1 7.3
An alternative is to use sfdisfil.
bash$ sfdisfil > file.asc col=3 format="%3.1f " number=n < file.rsf bash$ cat file.asc 1.0 1.5 3.0 4.8 9.1 7.3
Reading and writing CSV files[edit]
CSV (Comma-separated values) is a particular example of an ASCII format, where commas separate values on different rows or other symbols. To convert from CSV to RSF, you can use the sfcsv2rsf utility. For example, let us take an ASCII file with numbers separated by commas
bash$ cat file.csv 1.0,1.5,3.0 4.8,9.1,7.3
Converting it to RSF:
bash$ sfcsv2rsf < file.csv > file.rsf
bash$ sfin file.rsf
file.rsf:
in="/tmp/file.rsf@"
esize=4 type=float form=native
n1=3 d1=1 o1=0 label1="unknown" unit1="unknown"
n2=2 d2=1 o2=0 label2="unknown" unit2="unknown"
6 elements 24 bytes
To convert from RSF to CSV, we can use formatting parameters in sfdd:
bash$ sfdd form=ascii --out=file.csv \ line=3 strip=1 format="%3.1f," < file.rsf >/dev/null bash$ cat file.csv 1.0,1.5,3.0 4.8,9.1,7.3
Some CSV files contain headers with definitions for different columns.
bash$ cat file.csv height,width,weight 1.0,1.5,3.0 4.8,9.1,7.3
To read a file like that, use header= parameter in sfcsv2rsf, as follows:
bash$ sfcsv2rsf < file.csv header=y > file.rsf
After that, different columns can be accessed by keywords.
bash$ < file.rsf sfheaderattr segy=n
3 headers, 2 traces
*******************************************************************************
key min max mean
-------------------------------------------------------------------------------
height 0 1 @ 0 4.8 @ 1 2.9
width 1 1.5 @ 0 9.1 @ 1 5.3
weight 2 3 @ 0 7.3 @ 1 5.15
*******************************************************************************
Reading LAS files[edit]
LAS (Log ASCII Standard) is a text format used for storing well-logging data (Heslop et al., 1999[8]). LAS files can be converted to the RSF format using sflas2rsf utility. Let us try an example file from one of the SEG tutorials:
bash$ tutorials=https://raw.githubusercontent.com/seg/tutorials-2014/master bash$ wget $tutorials/1406_Make_a_synthetic/L-30.las
Converting to RSF, we can detect 15 different logs:
bash$ sflas2rsf L-30.las L-30.rsf
(base) sergey@DESKTOP-80QRDA0:~/all/fomels/nnint$ sfin L-30.rsf
L-30.rsf:
in="/home/sergey/RSFROOT/data/L-30.rsf@"
esize=4 type=float form=native
n1=15 d1=? o1=?
n2=25621 d2=0.5 o2=1140
384315 elements 1537260 bytes
Individual logs are accessible by their keys and can be used in programs like sfheadermath.
bash$ < L-30.rsf sfheaderattr segy=n desc=y
15 headers, 25621 traces
*******************************************************************************
key min max mean
-------------------------------------------------------------------------------
DEPTH 0 1140 @ 0 13950 @ 25620 7545
[Depth]
CALD 1 -999 @ 0 19.811 @ 3909 -140.356
[Caliper Caliper - Density]
CALS 2 -999 @ 0 14.84 @ 23096 7.43849
[Caliper Caliper - Sonic]
DEPT 3 1140 @ 0 13950 @ 25620 7545
[Depth]
DRHO 4 -999 @ 0 0.254 @ 23667 -149.67
[Drho Delta Rho]
DT 5 -999 @ 0 199.263 @ 1462 90.0167
[Sonic Delta-T]
GRD 6 -999 @ 0 178.416 @ 21788 -100.952
[GammaRay Gamma Ray - Density]
GRS 7 -999 @ 0 140.148 @ 23376 53.8002
[GammaRay Gamma Ray - Sonic]
ILD 8 -999 @ 0 2022.95 @ 20 34.5917
[DeepRes Deep Induction Standard Processed Resistivity]
ILM 9 -999 @ 0 2196.26 @ 20661 40.5595
[MedRes Medium Induction Standard Processed Resistivity]
LL8 10 -999 @ 0 2097.76 @ 20213 35.6343
[ShalRes Latero-Log 8]
NPHILS 11 -999 @ 0 0.45 @ 23039 -776.522
[Neutron Neutron Porosity - Ls Mtx]
NPHISS 12 -999 @ 0 0.615 @ 5215 -373.244
[Neutron Neutron Porosity - Ss Mtx]
RHOB 13 -999 @ 0 2.811 @ 23941 -147.773
[Density Bulk Density]
SP 14 -999 @ 0 -19.065 @ 20570 -105.029
[SP Spontaneous Potential]
*******************************************************************************
bash$ < L-30.rsf sfheadermath output=RHOB segy=n > RHOB.rsf
bash$ < RHOB.rsf sfwindow min2=4000 max2=13000 | sfgraph title=Density
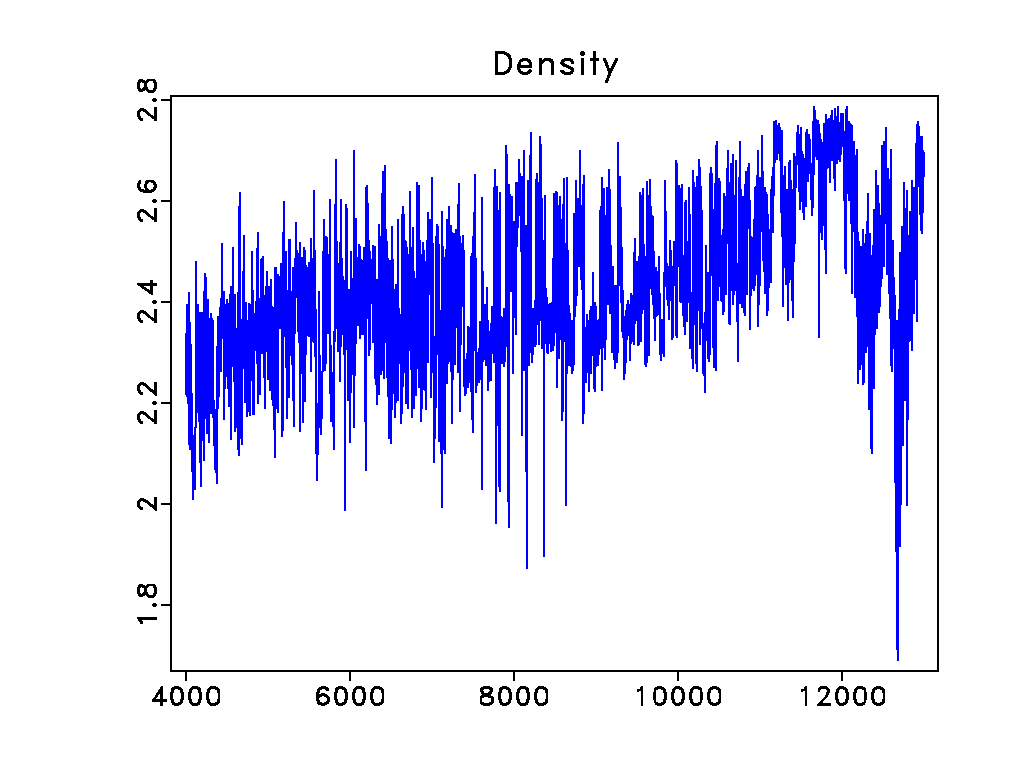
Other documentation[edit]
This note should give you a general understanding of the RSF file format. See the RSF Comprehensive Description if you want minutia. Other relevant documentation is:
- Why Madagascar
- Installation instructions
- Madagascar self-documentation
- Guide to madagascar programs
- Guide to the Madagascar programming interface
- Guide to programming with madagascar
- Revisiting SEP tour with Madagascar and SCons
- Reproducible computational experiments using SCons
About this document[edit]
This page was created from the LaTeX source in book/rsf/rsf/format.tex using latex2wiki.
References[edit]
- ↑ Claerbout, J. F., 1991, Introduction to Seplib and SEP utility software, in SEP-70, 413--436. Stanford Exploration Project.
- ↑ Raymond, E. S., 2004, The art of UNIX programming: Addison-Wesley.
- ↑ Biondi, B., R. Clapp, and S. Crawley, 1996, SEPlib90: SEPlib for 3-D prestack data, in SEP-92, 343--364. Stanford Exploration Project.
- ↑ Barry, K. M., D. A. Cavers, and C. W. Kneale, 1975, Report on recommended standards for digital tape formats: Geophysics, 40, 344--352
- ↑ See http://www.seg.org/SEGportalWEBproject/prod/SEG-Publications/Pub-Technical-Standards/Documents/seg_y_rev1.pdf
- ↑ Stockwell, J. W., 1997, Free software in education: A case study of CWP/SU: Seismic Unix: The Leading Edge, 16, 1045--1049.
- ↑ -------- 1999, The CWP/SU: Seismic Un*x package: Computers and Geosciences, 25, 415--419.
- ↑ Heslop, K., J. Karst, S. Prensky, D. Schmitt, et al., 1999, Log ASCII standard LAS version 3.0: The Log Analyst, 40.
