Reproducible computational experiments using SCons: Difference between revisions
typos |
|||
| (5 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
SCons (from Software Construction) is a well-known open-source | SCons (from Software Construction) is a well-known open-source | ||
program designed primarily for building software. | program designed primarily for building software. This paper describes our method of extending SCons for managing data processing | ||
flows and reproducible computational experiments. We demonstrate our | flows and reproducible computational experiments. We demonstrate our | ||
usage of SCons with a couple of simple examples. | usage of SCons with a couple of simple examples. | ||
| Line 11: | Line 10: | ||
experiments developed as part of the "Madagascar" software package. | experiments developed as part of the "Madagascar" software package. | ||
To reproduce the example experiments in this paper, you can download | To reproduce the example experiments in this paper, you can download | ||
Madagascar from | Madagascar from https://www.ahay.org . At the moment, the | ||
main Madagascar interface is the Unix shell command line so that you | main Madagascar interface is the Unix shell command line so that you | ||
will need a Unix/POSIX system (Linux, Mac OS X, Solaris, etc.) or Unix | will need a Unix/POSIX system (Linux, Mac OS X, Solaris, etc.) or Unix | ||
emulation under Windows (Cygwin, SFU, etc.) | emulation under Windows (Cygwin, SFU, etc.) | ||
Our focus, however, is not only on particular | Our focus, however, is not only on particular tools we use in our research but also on the general philosophy of | ||
tools we use in our research but also on the general philosophy of | |||
reproducible computations. | reproducible computations. | ||
===Reproducible research philosophy=== | ===Reproducible research philosophy=== | ||
Peer review is the backbone of scientific progress. From the ancient | Peer review is the backbone of scientific progress. From the ancient | ||
alchemists | alchemists who worked secretly on magic solutions to insolvable | ||
problems, | problems, modern science has come a long way to become a social | ||
enterprise | enterprise where the community openly publishes and verifies hypotheses, theories, and experimental results. By reproducing and | ||
verifying previously published research, a researcher can take new | verifying previously published research, a researcher can take new | ||
steps to advance the progress of science. | steps to advance the progress of science. | ||
Traditionally, scientific disciplines are divided into theoretical and | Traditionally, scientific disciplines are divided into theoretical and | ||
experimental studies. | experimental studies. The reproduction and verification of theoretical | ||
results usually | results usually require only imagination (apart from pencils and | ||
paper), experimental results are verified in laboratories using | paper), and experimental results are verified in laboratories using | ||
equipment and materials similar to those described in the publication. | equipment and materials similar to those described in the publication. | ||
During the last century, computational studies emerged as a new | During the last century, computational studies emerged as a new | ||
| Line 35: | Line 32: | ||
computer by applying numerical algorithms to digital data. How | computer by applying numerical algorithms to digital data. How | ||
reproducible are such experiments? On one hand, reproducing the result | reproducible are such experiments? On one hand, reproducing the result | ||
of a numerical experiment is | of a numerical experiment is difficult. The reader needs | ||
to have access to precisely the same kind of input data, software and | to have access to precisely the same kind of input data, software, and | ||
hardware as the author | hardware as the publication's author to reproduce the | ||
published result. It is often difficult or impossible to provide | published result. It is often difficult or impossible to provide | ||
detailed specifications for these components. On the other hand, | detailed specifications for these components. On the other hand, essential | ||
computational system components such as operating systems and | computational system components such as operating systems and | ||
file formats are getting increasingly standardized | file formats are getting increasingly standardized. New components | ||
can be shared in principle because they | can be shared in principle because they represent digital | ||
information transferable over the Internet. | information transferable over the Internet. | ||
The practice of software sharing has fueled the miraculously efficient | The practice of software sharing has fueled the miraculously efficient | ||
development of Linux, Apache, and many other open-source software | development of Linux, Apache, and many other open-source software | ||
projects. | projects. Its proponents often refer to this ideology as an analog of | ||
the scientific peer review tradition. Eric Raymond, a well-known | the scientific peer review tradition. Eric Raymond, a well-known | ||
open-source advocate | open-source advocate writes (Raymond, 2004<ref>Raymond, E. S., 2004, The art of UNIX programming: Addison-Wesley.</ref>): | ||
<blockquote> | <blockquote> | ||
Abandoning the habit of secrecy in favor of process transparency and | Abandoning the habit of secrecy in favor of process transparency and | ||
| Line 56: | Line 53: | ||
development as a discipline. | development as a discipline. | ||
</blockquote> | </blockquote> | ||
While software development | While software development tries to imitate science, computational | ||
science | science must borrow from the open-source model to sustain | ||
itself as a fully scientific discipline. In words of Randy LeVeque, a | itself as a fully scientific discipline. In the words of Randy LeVeque, a | ||
prominent mathematician (LeVeque, 2006<ref>LeVeque, R. J., to appear, 2006, Wave propagation software, computational | prominent mathematician (LeVeque, 2006<ref>LeVeque, R. J., to appear, 2006, Wave propagation software, computational science, and reproducible research: Presented at the Proc. International Congress of Mathematicians.</ref>), | ||
<blockquote> | <blockquote> | ||
Within the world of science, computation is now rightly seen as a | Within the world of science, computation is now rightly seen as a | ||
third vertex of a triangle complementing experiment and | third vertex of a triangle, complementing experiment and | ||
theory. However, as it is now often practiced, one can make a good | theory. However, as it is now often practiced, one can make a good case that computing is the last refuge of the scientific scoundrel | ||
case that computing is the last refuge of the scientific scoundrel | |||
[...] Where else in science can one get away with publishing | [...] Where else in science can one get away with publishing | ||
observations that are claimed to prove a theory or illustrate the | observations that are claimed to prove a theory or illustrate the | ||
success of a technique without having to give a careful description of | success of a technique without having to give a careful description of | ||
the methods used | the methods used in sufficient detail that others can attempt to | ||
repeat the experiment? [...] Scientific and mathematical journals are | repeat the experiment? [...] Scientific and mathematical journals are | ||
filled with pretty pictures these days of computational experiments | filled with pretty pictures these days of computational experiments | ||
that the reader has no hope of repeating. Even brilliant and well | that the reader has no hope of repeating. Even brilliant and well-intentioned computational scientists often do a poor job of presenting | ||
intentioned computational scientists often do a poor job of presenting | |||
their work in a reproducible manner. The methods are often very | their work in a reproducible manner. The methods are often very | ||
vaguely defined, and even if they are carefully defined, they would | vaguely defined, and even if they are carefully defined, they would | ||
| Line 78: | Line 73: | ||
test them. | test them. | ||
</blockquote> | </blockquote> | ||
In computer science, the concept of publishing and explaining computer | In computer science, the concept of publishing and explaining computer programs goes back to the idea of ''literate programming'' promoted | ||
programs goes back to the idea of ''literate programming'' promoted | |||
by Knuth (1984<ref>Knuth, D. E., 1984, Literate programming: Computer Journal, '''27''', 97--111.</ref>) and expended by many other researchers | by Knuth (1984<ref>Knuth, D. E., 1984, Literate programming: Computer Journal, '''27''', 97--111.</ref>) and expended by many other researchers | ||
(Thimbleby, 2003<ref>Thimbleby, H., 2003, Explaining code for publication: Software - Practice & Experience, '''33''', 975--908.</ref>). In his 2004 lecture on " | (Thimbleby, 2003<ref>Thimbleby, H., 2003, Explaining code for publication: Software - Practice & Experience, '''33''', 975--908.</ref>). In his 2004 lecture on "Better Programming," | ||
Harold Thimbleby notes<ref>http://www.uclic.ucl.ac.uk/harold/</ref> | Harold Thimbleby notes<ref>http://www.uclic.ucl.ac.uk/harold/</ref> | ||
<blockquote> | <blockquote> | ||
| Line 91: | Line 85: | ||
</blockquote> | </blockquote> | ||
<!-- | <!-- | ||
The quest for peer review and reproducibility is | The quest for peer review and reproducibility is vital | ||
for computational geosciences and computational geophysics in | for computational geosciences and computational geophysics in | ||
particular. The very first paper published in ''Geophysics'' was | particular. The very first paper published in ''Geophysics'' was | ||
titled "Black | titled "Black Magic in Geophysical Prospecting" | ||
() and presented an account | () and presented an account | ||
of different "magical" methods of oil explorations promoted by | of different "magical" methods of oil explorations promoted by | ||
entrepreneurs in the early days of geophysical exploration industry. | entrepreneurs in the early days of the geophysical exploration industry. | ||
Although none of these methods exist today, it is not a secret that | Although none of these methods exist today, it is not a secret that | ||
industrial practice is full of nearly magical tricks, often hidden | industrial practice is full of nearly magical tricks, often hidden | ||
| Line 106: | Line 100: | ||
Nearly ten years ago, the technology of reproducible research in | Nearly ten years ago, the technology of reproducible research in | ||
geophysics was pioneered by Jon Claerbout and his students at the | geophysics was pioneered by Jon Claerbout and his students at the | ||
Stanford Exploration Project (SEP). | Stanford Exploration Project (SEP). SEP's system of reproducible | ||
research requires the author of a publication to document creation of | research requires the author of a publication to document the creation of | ||
numerical results from the input data and software sources to let | numerical results from the input data and software sources to let | ||
others test and verify the | others test and verify the reproducibility of the results | ||
(Claerbout, 1992a<ref>Claerbout, J., 1992a, Electronic documents give reproducible research a new | (Claerbout, 1992a<ref>Claerbout, J., 1992a, Electronic documents give reproducible research a new meaning: 62nd Ann. Internat. Mtg, 601--604, Soc. of Expl. Geophys.</ref>;Schwab et al., 2000<ref>Schwab, M., M. Karrenbach, and J. Claerbout, 2000, Making scientific computations reproducible: Computing in Science & Engineering, '''2''', 61--67.</ref>). | ||
The discipline of reproducible research was also adopted and | The discipline of reproducible research was also adopted and | ||
popularized in the statistics and wavelet theory community by | popularized in the statistics and wavelet theory community by | ||
Buckheit and Donoho (1995<ref>Buckheit, J. and D. L. Donoho, 1995, Wavelab and reproducible research, ''in'' Wavelets and Statistics, volume '''103''', 55--81. Springer-Verlag.</ref>). It is referenced in several popular wavelet theory | Buckheit and Donoho (1995<ref>Buckheit, J. and D. L. Donoho, 1995, Wavelab and reproducible research, ''in'' Wavelets and Statistics, volume '''103''', 55--81. Springer-Verlag.</ref>). It is referenced in several popular wavelet theory | ||
books (Hubbard, 1998<ref>Hubbard, B. B., 1998, The world according to wavelets: The story of a | books (Hubbard, 1998<ref>Hubbard, B. B., 1998, The world according to wavelets: The story of a mathematical technique in the making: AK Peters.</ref>;Mallat, 1999<ref>Mallat, S., 1999, A wavelet tour of signal processing: Academic Press.</ref>). Pledges for reproducible research | ||
appear nowadays in fields as diverse as | appear nowadays in fields as diverse as | ||
bioinformatics | bioinformatics | ||
(Gentleman et al., 2004<ref>Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang, 2004, Bioconductor: open software development for computational biology and | (Gentleman et al., 2004<ref>Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang, 2004, Bioconductor: open software development for computational biology and bioinformatics: Genome Biology, '''5''', R80.</ref>), | ||
geoinformatics (Bivand, 2006<ref>Bivand, R., 2006, Implementing spatial data analysis software tools in r: Geographical Analysis, '''38''', 23--40.</ref>), and computational | geoinformatics (Bivand, 2006<ref>Bivand, R., 2006, Implementing spatial data analysis software tools in r: Geographical Analysis, '''38''', 23--40.</ref>), and computational wave propagation (LeVeque, 2006<ref>LeVeque, R. J., to appear, 2006, Wave propagation software, computational science, and reproducible research: Presented at the Proc. International Congress of Mathematicians.</ref>). However, computational scientists' adoption of reproducible research practice has been slow. | ||
wave propagation (LeVeque, 2006<ref>LeVeque, R. J., to appear, 2006, Wave propagation software, computational | Partially, this is caused by complicated and inadequate tools. | ||
research practice | |||
Partially, this is caused by | |||
===Tools for reproducible research=== | ===Tools for reproducible research=== | ||
The reproducible research system developed at Stanford is based on | The reproducible research system developed at Stanford is based on | ||
"make" (Stallman et al., 2004<ref>Stallman, R. M., R. McGrath, and P. D. Smith, 2004, GNU make: A program | "make" (Stallman et al., 2004<ref>Stallman, R. M., R. McGrath, and P. D. Smith, 2004, GNU make: A program for directing recompilation: GNU Press.</ref>), a Unix software construction utility. | ||
Initially, SEP used "cake," a dialect of "make" | |||
(Nichols and Cole, 1989<ref>Nichols, D. and S. Cole, 1989, Device independent software installation with CAKE, ''in'' SEP-61, 341--344. Stanford Exploration Project.</ref>;Claerbout and Nichols, 1990<ref>Claerbout, J. F. and D. Nichols, 1990, Why active documents need cake, ''in'' SEP-67, 145--148. Stanford Exploration Project.</ref>;Claerbout, 1992b<ref>-------- 1992b, How to use Cake with interactive documents, ''in'' SEP-73, 451--460. Stanford Exploration Project.</ref>;Claerbout and Karrenbach, 1993<ref>Claerbout, J. F. and M. Karrenbach, 1993, How to use cake with interactive documents, ''in'' SEP-77, 427--444. Stanford Exploration Project.</ref>). | (Nichols and Cole, 1989<ref>Nichols, D. and S. Cole, 1989, Device independent software installation with CAKE, ''in'' SEP-61, 341--344. Stanford Exploration Project.</ref>;Claerbout and Nichols, 1990<ref>Claerbout, J. F. and D. Nichols, 1990, Why active documents need cake, ''in'' SEP-67, 145--148. Stanford Exploration Project.</ref>;Claerbout, 1992b<ref>-------- 1992b, How to use Cake with interactive documents, ''in'' SEP-73, 451--460. Stanford Exploration Project.</ref>;Claerbout and Karrenbach, 1993<ref>Claerbout, J. F. and M. Karrenbach, 1993, How to use cake with interactive documents, ''in'' SEP-77, 427--444. Stanford Exploration Project.</ref>). | ||
The system was converted to "GNU make" | The system was converted to "GNU make," a more standard dialect, by | ||
Schwab and Schroeder (1995<ref>Schwab, M. and J. Schroeder, 1995, Reproducible research documents using GNUmake, ''in'' SEP-89, 217--226. Stanford Exploration Project.</ref>). The | Schwab and Schroeder (1995<ref>Schwab, M. and J. Schroeder, 1995, Reproducible research documents using GNUmake, ''in'' SEP-89, 217--226. Stanford Exploration Project.</ref>). The "make" program keeps track of dependencies between different | ||
"make" program keeps track of dependencies between different | |||
components of the system and the software construction targets, which, | components of the system and the software construction targets, which, | ||
in the case of a reproducible research system, turn into figures and | in the case of a reproducible research system, turn into figures and | ||
manuscripts. The targets and commands for their construction | manuscripts. The author specifies the targets and commands for their construction in "makefiles," which serve as databases for | ||
defining source and target dependencies. A dependency-based system | defining source and target dependencies. A dependency-based system | ||
leads to rapid development | leads to rapid development because when one of the sources changes, | ||
only parts that depend on this source get recomputed. | only parts that depend on this source get recomputed. Buckheit and Donoho (1995<ref>Buckheit, J. and D. L. Donoho, 1995, Wavelab and reproducible research, ''in'' Wavelets and Statistics, volume '''103''', 55--81. Springer-Verlag.</ref>) | ||
based their system on MATLAB, a popular integrated development | based their system on MATLAB, a popular integrated development | ||
environment produced by MathWorks (Sigmon and Davis, 2001<ref>Sigmon, K. and T. A. Davis, 2001, MATLAB primer, sixth edition: Chapman & Hall.</ref>). | environment produced by MathWorks (Sigmon and Davis, 2001<ref>Sigmon, K. and T. A. Davis, 2001, MATLAB primer, sixth edition: Chapman & Hall.</ref>). While MATLAB is an adequate tool for prototyping numerical algorithms, it may not be | ||
adequate tool for prototyping numerical algorithms, it may not be | |||
sufficient for large-scale computations typical for many applications | sufficient for large-scale computations typical for many applications | ||
in computational geophysics. | in computational geophysics. | ||
"Make" is | "Make" is a handy utility employed by thousands of | ||
software development projects. Unfortunately, it is not | software development projects. Unfortunately, it is not | ||
well designed from the user experience | well designed from the perspective of user experience. "Make" employs | ||
an obscure and limited special language (a mixture of Unix shell | an obscure and limited special language (a mixture of Unix shell | ||
and special-purpose commands), which often appears confusing | |||
to | to inexperienced users. According to Peter van der Linden, a software | ||
expert from Sun Microsystems (van der Linden, 1994<ref>van der Linden, P., 1994, Expert C programming: Prentice Hall.</ref>), | expert from Sun Microsystems (van der Linden, 1994<ref>van der Linden, P., 1994, Expert C programming: Prentice Hall.</ref>), | ||
<blockquote> | <blockquote> | ||
"Sendmail" and "make" are two well known programs that are | "Sendmail" and "make" are two well-known programs that are | ||
pretty widely regarded as originally being debugged into existence. | pretty widely regarded as originally being debugged into existence. | ||
That's why their command languages are so poorly thought out and | That's why their command languages are so poorly thought out and | ||
| Line 158: | Line 147: | ||
troublesome. | troublesome. | ||
</blockquote> | </blockquote> | ||
The inconvenience of "make" command language is also in its limited | The inconvenience of the "make" command language is also in its limited | ||
capabilities. | capabilities. The reproducible research system developed by | ||
Schwab et al. (2000<ref>Schwab, M., M. Karrenbach, and J. Claerbout, 2000, Making scientific | Schwab et al. (2000<ref>Schwab, M., M. Karrenbach, and J. Claerbout, 2000, Making scientific computations reproducible: Computing in Science & Engineering, '''2''', 61--67.</ref>) includes not only custom "make" rules but also an obscure and hardly portable agglomeration of shell and Perl scripts that extend "make" (Fomel et al., 1997<ref>Fomel, S., M. Schwab, and J. Schroeder, 1997, Empowering SEP's documents, ''in'' SEP-94, 339--361. Stanford Exploration Project.</ref>). | ||
obscure and hardly portable agglomeration of shell and Perl scripts | |||
that extend "make" (Fomel et al., 1997<ref>Fomel, S., M. Schwab, and J. Schroeder, 1997, Empowering SEP's documents, ''in'' SEP-94, 339--361. Stanford Exploration Project.</ref>). | |||
Several alternative systems for dependency-checking software | Several alternative systems for dependency-checking software | ||
construction have been developed in recent years. One of the most | construction have been developed in recent years. One of the most | ||
promising new tools is SCons, enthusiastically endorsed by | promising new tools is SCons, enthusiastically endorsed by | ||
Dubois (2003<ref>Dubois, P. F., 2003, Why Johnny can't build: Computing in Science & Engineering, '''5''', 83--88.</ref>). The SCons initial design won the Software Carpentry | Dubois (2003<ref>Dubois, P. F., 2003, Why Johnny can't build: Computing in Science & Engineering, '''5''', 83--88.</ref>). The SCons initial design won the Software Carpentry competition sponsored by Los Alamos National Laboratory in 2000 in the category of "a dependency management tool to replace make." Some of the main advantages of SCons are: | ||
competition sponsored by Los Alamos National Laboratory in 2000 in the | |||
category of "a dependency management tool to replace make" | |||
the main advantages of SCons are: | |||
*SCons configuration files are Python scripts. Python is a modern programming language praised for its readability, elegance, simplicity, and power (Rossum, 2000a<ref>Rossum, G. V., 2000a, Python reference manual: Iuniverse Inc.</ref>;Rossum, 2000b<ref>-------- 2000b, Python tutorial: Iuniverse Inc.</ref>). Scales and Ecke (2002<ref>Scales, J. A. and H. Ecke, 2002, What programming languages should we teach | *SCons configuration files are Python scripts. Python is a modern programming language praised for its readability, elegance, simplicity, and power (Rossum, 2000a<ref>Rossum, G. V., 2000a, Python reference manual: Iuniverse Inc.</ref>;Rossum, 2000b<ref>-------- 2000b, Python tutorial: Iuniverse Inc.</ref>). Scales and Ecke (2002<ref>Scales, J. A. and H. Ecke, 2002, What programming languages should we teach our undergraduates?: The Leading Edge, '''21''', 260--267.</ref>) recommend Python as the first programming language for geophysics students. | ||
*SCons offers reliable, automatic, and extensible dependency analysis and creates a global view of all | *SCons offers reliable, automatic, and extensible dependency analysis and creates a global view of all dependencies—no more "make depend," "make clean," or multiple build passes of touching and reordering targets to get all the dependencies. | ||
*SCons has built-in support for many programming languages and systems | *SCons has built-in support for many programming languages and systems, including C, C++, Fortran, Java, and LaTeX. | ||
*While "make" relies on timestamps | *While "make" relies on timestamps to detect file changes (creating numerous problems on platforms with different system clocks), SCons uses a more reliable detection mechanism, employing MD5 signatures by default. It can detect changes not only in files but also in commands used to build them. | ||
*SCons provides integrated support for parallel builds. | *SCons provides integrated support for parallel builds. | ||
*SCons provides configuration support analogous to the "autoconf" utility for testing the environment on different platforms. | *SCons provides configuration support analogous to the "autoconf" utility for testing the environment on different platforms. | ||
*SCons is designed from the ground up as a cross-platform tool. It | *SCons is designed from the ground up as a cross-platform tool. It works equally well on POSIX systems (Linux, Mac OS X, Solaris, etc.) and Windows. | ||
*The stability of SCons is assured by an incremental development methodology utilizing comprehensive regression tests. | *The stability of SCons is assured by an incremental development methodology utilizing comprehensive regression tests. | ||
*SCons is publicly released under a liberal open-source license<ref>As | *SCons is publicly released under a liberal open-source license<ref>As of this writing, SCons is in a beta version of 0.96, approaching the 1.0 official release. See http://www.scons.org/.</ref>. | ||
In this paper, we propose to adopt SCons as a new platform for | In this paper, we propose to adopt SCons as a new platform for | ||
reproducible research in scientific computing. | reproducible research in scientific computing. | ||
===Paper organization=== | ===Paper organization=== | ||
To demonstrate our adoption of SCons for reproducible research, we | To demonstrate our adoption of SCons for reproducible research, we first describe a couple of simple examples of computational | ||
first describe a couple of simple examples of computational | |||
experiments and then show how SCons helps us document our | experiments and then show how SCons helps us document our | ||
computational results. | computational results. | ||
<!-- | <!-- | ||
\newpage | \newpage | ||
==Madagascar open source code== | ==Madagascar open-source code== | ||
Madagascar's homepage is http://rsf.sourceforge.net. Madagascar | Madagascar's homepage is http://rsf.sourceforge.net. Madagascar | ||
| Line 200: | Line 183: | ||
installed on different platforms and tested before being released. | installed on different platforms and tested before being released. | ||
Updates are typically done every few months as opposed to the | Updates are typically done every few months as opposed to the | ||
development version which is updated every few hours or days by a | development version, which is updated every few hours or days by a | ||
dynamic team of developers. As such, there is no guarantee that the | dynamic team of developers. As such, there is no guarantee that the | ||
development version will be fully functional and stable at any given | development version will be fully functional and stable at any given | ||
time. In the remainder of this paper, we assume that you have | |||
successfully installed Madagascar stable version and that you have an | successfully installed Madagascar stable version and that you have an | ||
Internet connection\footnote{XXX provide alternate | Internet connection\footnote{XXX provide alternate means to download | ||
Lena.img if no Internet connection XXX}. | Lena.img if no Internet connection XXX}. | ||
--> | --> | ||
| Line 211: | Line 194: | ||
==Example experiments== | ==Example experiments== | ||
The main <tt>SConstruct</tt> commands defined in our reproducible | The main <tt>SConstruct</tt> commands defined in our reproducible research environment are collected in the table. | ||
research environment are collected in the table. | |||
<center> | <center> | ||
| Line 252: | Line 234: | ||
To follow the first example, select a working project directory and | To follow the first example, select a working project directory and | ||
copy the following code | copy the following code | ||
to a file named <tt>SConstruct</tt><ref>The source of this file is also accessible at [http://sourceforge.net/p/rsf/code/HEAD/tree/trunk/book/rsf/scons/easystart/SConstruct book/rsf/scons/easystart/SConstruct].</ref>. | to a file named <tt>SConstruct</tt><ref>The source of this file is also accessible at [http://sourceforge.net/p/rsf/code/HEAD/tree/trunk/book/rsf/scons/easystart/SConstruct $RSFSRC/book/rsf/scons/easystart/SConstruct].</ref>. | ||
<python> | <syntaxhighlight lang="python"> | ||
from rsf.proj import * | from rsf.proj import * | ||
| Line 265: | Line 247: | ||
stdin=0) | stdin=0) | ||
# Convert to floating point and window out first trace | # Convert to floating point and window out the first trace | ||
Flow('lena','lena.hdr','dd type=float | window f2=1') | Flow('lena','lena.hdr','dd type=float | window f2=1') | ||
| Line 277: | Line 259: | ||
# Wrap up | # Wrap up | ||
End() | End() | ||
</ | </syntaxhighlight> | ||
This is our "hello | This is our "hello world" example that illustrates the basic use of | ||
some of the commands presented in Table~(tbl:commands). The plan | some of the commands presented in Table~(tbl:commands). The plan | ||
for this experiment is | for this experiment is to download data from a public data | ||
server, | server, convert it to an appropriate file format, and generate a | ||
figure for publication. But let us | figure for publication. But let us look at the | ||
<tt>SConstruct</tt> script and try to decorticate it. | <tt>SConstruct</tt> script and try to decorticate it. | ||
<python> | <syntaxhighlight lang="python"> | ||
from rsf.proj import * | from rsf.proj import * | ||
</ | </syntaxhighlight> | ||
| Line 298: | Line 280: | ||
<python> | <syntaxhighlight lang="python"> | ||
Fetch('lena.img','imgs') | Fetch('lena.img','imgs') | ||
</ | </syntaxhighlight> | ||
| Line 329: | Line 311: | ||
In the following examples, we will use <tt>-Q</tt> (quiet) option of | In the following examples, we will use <tt>-Q</tt> (quiet) option of | ||
<tt>scons</tt> to suppress the verbose output. | <tt>scons</tt> to suppress the verbose output. | ||
<python> | <syntaxhighlight lang="python"> | ||
Flow('lena.hdr','lena.img', | Flow('lena.hdr','lena.img', | ||
'echo n1=512 n2=513 in=$SOURCE data_format=native_uchar', | 'echo n1=512 n2=513 in=$SOURCE data_format=native_uchar', | ||
stdin=0) | stdin=0) | ||
</ | </syntaxhighlight> | ||
| Line 345: | Line 327: | ||
Since <tt>echo</tt> does not take a standard input, stdin is set to 0 | Since <tt>echo</tt> does not take a standard input, stdin is set to 0 | ||
in the Flow command otherwise the first source is the standard input. | in the Flow command; otherwise, the first source is the standard input. | ||
Likewise, the first target is the standard output unless otherwise | Likewise, the first target is the standard output unless otherwise | ||
specified. | specified. | ||
| Line 352: | Line 334: | ||
Note that | Note that | ||
<tt>lena.img</tt> is referred as <tt>$SOURCE</tt> in the command. This | <tt>lena.img</tt> is referred as <tt>$SOURCE</tt> in the command. This | ||
allows us to change | allows us to change the source file's name without changing the command. | ||
The data format of the <tt>lena.img</tt> image file is <tt>uchar</tt> | The data format of the <tt>lena.img</tt> image file is <tt>uchar</tt> | ||
(unsigned character), the image consists of 513 traces with 512 | (unsigned character), the image consists of 513 traces with 512 | ||
samples per trace. | samples per trace. Our next step is to convert the image | ||
representation to floating point numbers and to window out the first | representation to floating point numbers and to window out the first | ||
trace so that the final image is | trace so that the final image is 512 by 512 square. The two | ||
transformations are conveniently combined into one with the help of a Unix pipe. | transformations are conveniently combined into one with the help of a Unix pipe. | ||
<python> | <syntaxhighlight lang="python"> | ||
Flow('lena','lena.hdr','dd type=float | window f2=1') | Flow('lena','lena.hdr','dd type=float | window f2=1') | ||
</ | </syntaxhighlight> | ||
<pre> | <pre> | ||
bash$ scons -Q lena | bash$ scons -Q lena | ||
scons: *** Do not know how to make target `lena'. | scons: *** Do not know how to make target `lena'. Stop. | ||
</pre> | </pre> | ||
What happened? In the absence of the file suffix, the <tt>Flow</tt> | What happened? In the absence of the file suffix, the <tt>Flow</tt> | ||
| Line 387: | Line 369: | ||
262144 elements 1048576 bytes | 262144 elements 1048576 bytes | ||
</pre> | </pre> | ||
In the last step, we will create a plot file | In the last step, we will create a plot file to display the image | ||
on the screen and for including it in the publication. | on the screen and for including it in the publication. | ||
<python> | <syntaxhighlight lang="python"> | ||
Result('lena', | Result('lena', | ||
''' | ''' | ||
| Line 396: | Line 378: | ||
clip=100 screenratio=1 | clip=100 screenratio=1 | ||
''') | ''') | ||
</ | </syntaxhighlight> | ||
Notice that we broke the long command string into multiple lines by | Notice that we broke the long command string into multiple lines by | ||
using Python's triple quote syntax. All the extra white space will be | using Python's triple quote syntax. All the extra white space will be | ||
ignored when the multiple line string gets translated into the command | ignored when the multiple-line string gets translated into the command | ||
line. | line. The <tt>Result</tt> command has special targets associated with | ||
it. Try, for example, "<tt>scons lena.view</tt>" to observe the | it. Try, for example, "<tt>scons lena.view</tt>" to observe the | ||
figure <tt>Fig/lena.vpl</tt> generated in a specially created | figure <tt>Fig/lena.vpl</tt> generated in a specially created | ||
| Line 412: | Line 394: | ||
The reproducible script ends with | The reproducible script ends with | ||
<python> | <syntaxhighlight lang="python"> | ||
End() | End() | ||
</ | </syntaxhighlight> | ||
Ready to experiment? Try some of the following: | Ready to experiment? Try some of the following: | ||
| Line 436: | Line 418: | ||
< lena.rsf /RSF/bin/sfgrey title="Hello, World!" transp=n color=b bias=128 clip=50 screenratio=1 > Fig/lena.vpl | < lena.rsf /RSF/bin/sfgrey title="Hello, World!" transp=n color=b bias=128 clip=50 screenratio=1 > Fig/lena.vpl | ||
sfpen Fig/lena.vpl | sfpen Fig/lena.vpl | ||
</pre> SCons is smart enough to recognize that your editing did not affect any of the previous results in the data flow chain! Keeping track of dependencies is the main feature that separates data processing and computational experimenting with SCons from using linear shell scripts. | </pre> SCons is smart enough to recognize that your editing did not affect any of the previous results in the data flow chain! Keeping track of dependencies is the main feature that separates data processing and computational experimenting with SCons from using linear shell scripts. This feature can save you a lot of time for computationally demanding data processing and make your experiments more interactive and enjoyable. | ||
#A special parameter to SCons (defined in <tt>rsfproj.py</tt>) can time the execution of each step in the processing flow. Try running <tt>scons TIMER=y</tt>. | #A special parameter to SCons (defined in <tt>rsfproj.py</tt>) can time the execution of each step in the processing flow. Try running <tt>scons TIMER=y</tt>. | ||
#The <tt>rsfproj</tt> module has direct access to the database that stores parameters of all Madagascar modules. Try running <tt>scons CHECKPAR=y</tt> to see parameter checking enforced before computations\footnote{This feature is new and experimental and may not work | #The <tt>rsfproj</tt> module has direct access to the database that stores the parameters of all Madagascar modules. Try running <tt>scons CHECKPAR=y</tt> to see parameter checking enforced before computations\footnote{This feature is new and experimental and may not work correctly yet}. | ||
The summary of our SCons commands is given in the table. | The summary of our SCons commands is given in the table. | ||
| Line 478: | Line 460: | ||
|style="background-color:#ffdead;"| '''<tt>scons CHECKPAR=y ...</tt> ''' | |style="background-color:#ffdead;"| '''<tt>scons CHECKPAR=y ...</tt> ''' | ||
|- | |- | ||
| Check the names and values of all parameters supplied to Madagascar modules in the processing | | Check the names and values of all parameters supplied to Madagascar modules in the processing flow before executing anything (guards against incorrect input.) This option is new and experimental. | ||
|} | |} | ||
| Line 484: | Line 466: | ||
The plan for this experiment is to add random noise to the test | The plan for this experiment is to add random noise to the test | ||
"Lena" image and then | "Lena" image and then attempt removing it by low-pass filtering | ||
and | and hard thresholding of coefficients in the Fourier domain. The | ||
resultant images are shown in the figures. | |||
[[Image:panel1.png|frame|center|Top left: original image. Top | [[Image:panel1.png|frame|center|Top left: original image. Top right: random noise added. Bottom left: original image spectrum in the Fourier (<math>F</math>-<math>X</math>) domain. Bottom right: noisy image spectrum in the Fourier (<math>F</math>-<math>X</math>) domain.]] | ||
[[Image:panel2.png|frame|center|Left: denoising by low-pass filtering. Right: denoising by hard thresholding in the Fourier domain.]] | [[Image:panel2.png|frame|center|Left: denoising by low-pass filtering. Right: denoising by hard thresholding in the Fourier domain.]] | ||
| Line 496: | Line 478: | ||
reproducible scripts. A demo script is available in the | reproducible scripts. A demo script is available in the | ||
<tt>rsf/scons/rsfpy</tt> subdirectory of the Madagascar <tt>book</tt> | <tt>rsf/scons/rsfpy</tt> subdirectory of the Madagascar <tt>book</tt> | ||
directory. Rather than commenting it line-by-line, we select some | directory. Rather than commenting on it line-by-line, we select some | ||
parts of interest. | parts of interest. | ||
In the <tt>SConstruct</tt> script, we can declare | In the <tt>SConstruct</tt> script, we can declare | ||
Python variables | Python variables | ||
<python> | <syntaxhighlight lang="python"> | ||
bias = 128 | bias = 128 | ||
</ | </syntaxhighlight> | ||
and use them later, for example, to define our customized plot | and use them later, for example, to define our customized plot | ||
command as a Python function | command as a Python function | ||
<python> | <syntaxhighlight lang="python"> | ||
def grey(title,transp='n',bias=bias): | def grey(title,transp='n',bias=bias): | ||
return ''' | return ''' | ||
| Line 513: | Line 495: | ||
label1= label2= | label1= label2= | ||
''' % (title,transp,bias) | ''' % (title,transp,bias) | ||
</ | </syntaxhighlight> | ||
This Python function, named <tt>grey()</tt>, can then be called in Plot or Result | This Python function, named <tt>grey()</tt>, can then be called in Plot or Result | ||
commands, e.g. | commands, e.g. | ||
<python> | <syntaxhighlight lang="python"> | ||
Plot('lplena',grey('Noisy Lena LP filtered')) | Plot('lplena',grey('Noisy Lena LP filtered')) | ||
</ | </syntaxhighlight> | ||
We can define a Python dictionary, e.g. | We can define a Python dictionary, e.g. | ||
<python> | <syntaxhighlight lang="python"> | ||
titles = {'lena':'Lena', | titles = {'lena':'Lena', | ||
'nlena':'Noisy Lena'} | 'nlena':'Noisy Lena'} | ||
</ | </syntaxhighlight> | ||
and loop over its entries, e.g. | and loop over its entries, e.g. | ||
<python> | <syntaxhighlight lang="python"> | ||
for name in titles.keys(): | for name in titles.keys(): | ||
Plot(name,grey(titles[name]) ) | Plot(name,grey(titles[name]) ) | ||
| Line 533: | Line 515: | ||
Flow('fx'+name,name,'sfspectra') | Flow('fx'+name,name,'sfspectra') | ||
Plot('fx'+name,grey(cftitle,'y',100)) | Plot('fx'+name,grey(cftitle,'y',100)) | ||
</ | </syntaxhighlight> | ||
Note that the title of the plots is obtained by concatenating Python | Note that the title of the plots is obtained by concatenating Python | ||
strings. | strings. | ||
Python strings can also be used to define sequences of commands used | Python strings can also be used to define sequences of commands used | ||
in several Flows, e.g. | in several Flows, e.g. | ||
<python> | <syntaxhighlight lang="python"> | ||
# 2-D FFT | # 2-D FFT | ||
fft2 = 'sffft1 sym=y | sffft3 sym=y' | fft2 = 'sffft1 sym=y | sffft3 sym=y' | ||
Flow('fnlena','nlena',fft2) | Flow('fnlena','nlena',fft2) | ||
</ | </syntaxhighlight> | ||
Finally, in our Madagascar reproducible script, we may want the option | Finally, in our Madagascar reproducible script, we may want the option | ||
to pass command line arguments when running SCons or use default | to pass command line arguments when running SCons or use default | ||
values otherwise, e.g. | values otherwise, e.g. | ||
<python> | <syntaxhighlight lang="python"> | ||
# denoising using thresholding in the Fourier domain | # denoising using thresholding in the Fourier domain | ||
fthr = float(ARGUMENTS.get('fthr', 70)) | fthr = float(ARGUMENTS.get('fthr', 70)) | ||
Flow('fthrlena','fnlena','sfthr thr=%f mode="hard"' % fthr) | Flow('fthrlena','fnlena','sfthr thr=%f mode="hard"' % fthr) | ||
</ | </syntaxhighlight> | ||
Running <tt>scons</tt> only, the default value set for fthr (i.e. 70) | Running <tt>scons</tt> only, the default value set for fthr (i.e. 70) | ||
is used whereas running <tt>scons fthr=68</tt> set fthr to a command | is used whereas running <tt>scons fthr=68</tt> set fthr to a command | ||
line specified value. | line specified value. | ||
This is by no | This is by no means an exhaustive list of options, but hopefully, it | ||
will give you a flavor of the powerful tool you have in your hands. Enjoy! | |||
<!-- | <!-- | ||
===Useful SCons commands for reproducible scripts=== | ===Useful SCons commands for reproducible scripts=== | ||
| Line 565: | Line 547: | ||
<tt>Fig</tt> folder) obtained in a Result command. <tt>scons view</tt> | <tt>Fig</tt> folder) obtained in a Result command. <tt>scons view</tt> | ||
displays the result plots one after the other. | displays the result plots one after the other. | ||
It is also possible to check parameters for Madagascar programs in | It is also possible to check the parameters for Madagascar programs in | ||
SCons Flow commands using the CHECKPAR option (\texttt{scons | SCons Flow commands using the CHECKPAR option (\texttt{scons | ||
CHECKPAR=y target}). Note that CHECKPAR is an experimental option | CHECKPAR=y target}). Note that CHECKPAR is an experimental option | ||
and will be enhanced in the future to include parameter ranges and | and will be enhanced in the future to include parameter ranges and | ||
other safety checks. | other safety checks. | ||
To time the execution of processing flows in a SConstruct use the | To time the execution of processing flows in a SConstruct, use the | ||
TIMER option (<tt>scons TIMER=y target</tt>). | TIMER option (<tt>scons TIMER=y target</tt>). | ||
<tt>scons lock</tt> is used to secure result plots and copy them from | <tt>scons lock</tt> is used to secure result plots and copy them from | ||
| Line 577: | Line 559: | ||
variable to the directory where you want Madagascar to put your key | variable to the directory where you want Madagascar to put your key | ||
Madagascar result plots. Note that this is a necessary step before | Madagascar result plots. Note that this is a necessary step before | ||
creating | creating reproducible documentation. <tt>scons plot.flip</tt> runs | ||
<tt>xtpen Fig/plot.vpl /locked/figures/plot.vpl</tt> to flip between | <tt>xtpen Fig/plot.vpl /locked/figures/plot.vpl</tt> to flip between | ||
the new and locked figure. This is useful when detecting changes. | the new and locked figure. This is useful when detecting changes. | ||
| Line 585: | Line 567: | ||
You are done with computational experiments and want to communicate | You are done with computational experiments and want to communicate | ||
them in a paper. SCons helps us create high-quality papers | them in a paper. SCons helps us create high-quality papers where | ||
computational results (figures) are integrated with papers written in | computational results (figures) are integrated with papers written in | ||
L<sup>A</sup>TEX\. | L<sup>A</sup>TEX\. | ||
| Line 591: | Line 573: | ||
<tt>RSFROOT</tt> is the environmental variable to the Madagascar | <tt>RSFROOT</tt> is the environmental variable to the Madagascar | ||
installation directory. The source of this file is in | installation directory. The source of this file is in | ||
[http:// | [http://sourceforge.net/p/rsf/code/HEAD/tree/trunk/framework/rsf/tex.py framework/rsf/tex.py]. | ||
We summarize the basic methods and commands in the tables. | We summarize the basic methods and commands in the tables. | ||
| Line 641: | Line 623: | ||
A Madagascar reproducible paper is a paper written in L<sup>A</sup>TEX and | A Madagascar reproducible paper is a paper written in L<sup>A</sup>TEX and | ||
whose figures are either generated by Madagascar reproducible scripts | whose figures are either generated by Madagascar reproducible scripts | ||
or available for download, e.g. | or available for download, e.g., this paper! (<tt>paper.tex</tt> | ||
available in the <tt>rsf/scons/</tt> directory of Madagascar book | available in the <tt>rsf/scons/</tt> directory of Madagascar book | ||
section). | section). | ||
| Line 657: | Line 639: | ||
in the directory above the projects directories. | in the directory above the projects directories. | ||
<python> | <syntaxhighlight lang="python"> | ||
from rsf.tex import * | from rsf.tex import * | ||
Paper('velan',use='hyperref,listings,color') | Paper('velan',use='hyperref,listings,color') | ||
End(use='hyperref,listings,color') | End(use='hyperref,listings,color') | ||
</syntaxhighlight> | |||
This <tt>SConstruct</tt> generates this paper, but it can also compile | |||
This <tt>SConstruct</tt> generates this paper but it can also compile | |||
<tt>velan.tex</tt> in the same directory. Note that there is no | <tt>velan.tex</tt> in the same directory. Note that there is no | ||
<tt>Paper</tt> command for <tt>paper.tex</tt> since it is the default | <tt>Paper</tt> command for <tt>paper.tex</tt> since it is the default | ||
| Line 671: | Line 652: | ||
<tt>paper.tex</tt> are passed in the End command. | <tt>paper.tex</tt> are passed in the End command. | ||
Let's now | Let's now take a closer look at <tt>paper.tex</tt> to understand how | ||
the figures of the documentation are linked to the reproducible | the figures of the documentation are linked to the reproducible | ||
scripts that created them. First of all, note that <tt>paper.tex</tt> | scripts that created them. First of all, note that <tt>paper.tex</tt> | ||
| Line 678: | Line 659: | ||
paper, the first figure was created in the project folder | paper, the first figure was created in the project folder | ||
<tt>easystart</tt> (sub-folder of our documentation folder) by the | <tt>easystart</tt> (sub-folder of our documentation folder) by the | ||
resulting plot <tt>lena.vpl</tt>. In the L<sup>A</sup>TEX source code, it | |||
translates as | translates as | ||
<latex> | <syntaxhighlight lang="latex"> | ||
\inputdir{easystart} | \inputdir{easystart} | ||
\sideplot{lena}{height=.25\textheight}{The output of the first numerical experiment.} | \sideplot{lena}{height=.25\textheight}{The output of the first numerical experiment.} | ||
</ | </syntaxhighlight> | ||
The <math>\backslash</math>inputdir command points to the project directory and | The <math>\backslash</math>inputdir command points to the project directory and | ||
| Line 690: | Line 671: | ||
L<sup>A</sup>TEX tag of the figure is <tt>fig:<math><</math>result_name<math>></math></tt>. The | L<sup>A</sup>TEX tag of the figure is <tt>fig:<math><</math>result_name<math>></math></tt>. The | ||
first time the paper is compiled, the result file is automatically | first time the paper is compiled, the result file is automatically | ||
converted to | converted to PDF format. | ||
<!-- | <!-- | ||
===Useful SCons commands for reproducible documentation=== | ===Useful SCons commands for reproducible documentation=== | ||
To | To compile this paper, you first need to run and lock the | ||
<tt>easystart</tt> project. Go in the <tt>easystart</tt> folder and | <tt>easystart</tt> project. Go in the <tt>easystart</tt> folder and | ||
run <tt>scons lock</tt>. Go back to the documentation folder and run | run <tt>scons lock</tt>. Go back to the documentation folder and run | ||
Latest revision as of 23:42, 20 November 2024
SCons (from Software Construction) is a well-known open-source program designed primarily for building software. This paper describes our method of extending SCons for managing data processing flows and reproducible computational experiments. We demonstrate our usage of SCons with a couple of simple examples.
Introduction[edit]
This paper introduces an environment for reproducible computational experiments developed as part of the "Madagascar" software package. To reproduce the example experiments in this paper, you can download Madagascar from https://www.ahay.org . At the moment, the main Madagascar interface is the Unix shell command line so that you will need a Unix/POSIX system (Linux, Mac OS X, Solaris, etc.) or Unix emulation under Windows (Cygwin, SFU, etc.) Our focus, however, is not only on particular tools we use in our research but also on the general philosophy of reproducible computations.
Reproducible research philosophy[edit]
Peer review is the backbone of scientific progress. From the ancient alchemists who worked secretly on magic solutions to insolvable problems, modern science has come a long way to become a social enterprise where the community openly publishes and verifies hypotheses, theories, and experimental results. By reproducing and verifying previously published research, a researcher can take new steps to advance the progress of science. Traditionally, scientific disciplines are divided into theoretical and experimental studies. The reproduction and verification of theoretical results usually require only imagination (apart from pencils and paper), and experimental results are verified in laboratories using equipment and materials similar to those described in the publication. During the last century, computational studies emerged as a new scientific discipline. Computational experiments are carried out on a computer by applying numerical algorithms to digital data. How reproducible are such experiments? On one hand, reproducing the result of a numerical experiment is difficult. The reader needs to have access to precisely the same kind of input data, software, and hardware as the publication's author to reproduce the published result. It is often difficult or impossible to provide detailed specifications for these components. On the other hand, essential computational system components such as operating systems and file formats are getting increasingly standardized. New components can be shared in principle because they represent digital information transferable over the Internet. The practice of software sharing has fueled the miraculously efficient development of Linux, Apache, and many other open-source software projects. Its proponents often refer to this ideology as an analog of the scientific peer review tradition. Eric Raymond, a well-known open-source advocate writes (Raymond, 2004[1]):
Abandoning the habit of secrecy in favor of process transparency and peer review was the crucial step by which alchemy became chemistry. In the same way, it is beginning to appear that open-source development may signal the long-awaited maturation of software development as a discipline.
While software development tries to imitate science, computational science must borrow from the open-source model to sustain itself as a fully scientific discipline. In the words of Randy LeVeque, a prominent mathematician (LeVeque, 2006[2]),
Within the world of science, computation is now rightly seen as a third vertex of a triangle, complementing experiment and theory. However, as it is now often practiced, one can make a good case that computing is the last refuge of the scientific scoundrel [...] Where else in science can one get away with publishing observations that are claimed to prove a theory or illustrate the success of a technique without having to give a careful description of the methods used in sufficient detail that others can attempt to repeat the experiment? [...] Scientific and mathematical journals are filled with pretty pictures these days of computational experiments that the reader has no hope of repeating. Even brilliant and well-intentioned computational scientists often do a poor job of presenting their work in a reproducible manner. The methods are often very vaguely defined, and even if they are carefully defined, they would normally have to be implemented from scratch by the reader in order to test them.
In computer science, the concept of publishing and explaining computer programs goes back to the idea of literate programming promoted by Knuth (1984[3]) and expended by many other researchers (Thimbleby, 2003[4]). In his 2004 lecture on "Better Programming," Harold Thimbleby notes[5]
We want ideas, and in particular programs, that work in one place to work elsewhere. One form of objectivity is that published science must work elsewhere than just in the author's laboratory or even just in the author's imagination; this requirement is called reproducibility .
Nearly ten years ago, the technology of reproducible research in geophysics was pioneered by Jon Claerbout and his students at the Stanford Exploration Project (SEP). SEP's system of reproducible research requires the author of a publication to document the creation of numerical results from the input data and software sources to let others test and verify the reproducibility of the results (Claerbout, 1992a[6];Schwab et al., 2000[7]).
The discipline of reproducible research was also adopted and popularized in the statistics and wavelet theory community by Buckheit and Donoho (1995[8]). It is referenced in several popular wavelet theory books (Hubbard, 1998[9];Mallat, 1999[10]). Pledges for reproducible research appear nowadays in fields as diverse as bioinformatics (Gentleman et al., 2004[11]), geoinformatics (Bivand, 2006[12]), and computational wave propagation (LeVeque, 2006[13]). However, computational scientists' adoption of reproducible research practice has been slow. Partially, this is caused by complicated and inadequate tools.
Tools for reproducible research[edit]
The reproducible research system developed at Stanford is based on "make" (Stallman et al., 2004[14]), a Unix software construction utility. Initially, SEP used "cake," a dialect of "make" (Nichols and Cole, 1989[15];Claerbout and Nichols, 1990[16];Claerbout, 1992b[17];Claerbout and Karrenbach, 1993[18]). The system was converted to "GNU make," a more standard dialect, by Schwab and Schroeder (1995[19]). The "make" program keeps track of dependencies between different components of the system and the software construction targets, which, in the case of a reproducible research system, turn into figures and manuscripts. The author specifies the targets and commands for their construction in "makefiles," which serve as databases for defining source and target dependencies. A dependency-based system leads to rapid development because when one of the sources changes, only parts that depend on this source get recomputed. Buckheit and Donoho (1995[20]) based their system on MATLAB, a popular integrated development environment produced by MathWorks (Sigmon and Davis, 2001[21]). While MATLAB is an adequate tool for prototyping numerical algorithms, it may not be sufficient for large-scale computations typical for many applications in computational geophysics. "Make" is a handy utility employed by thousands of software development projects. Unfortunately, it is not well designed from the perspective of user experience. "Make" employs an obscure and limited special language (a mixture of Unix shell and special-purpose commands), which often appears confusing to inexperienced users. According to Peter van der Linden, a software expert from Sun Microsystems (van der Linden, 1994[22]),
"Sendmail" and "make" are two well-known programs that are pretty widely regarded as originally being debugged into existence. That's why their command languages are so poorly thought out and difficult to learn. It's not just you -- everyone finds them troublesome.
The inconvenience of the "make" command language is also in its limited capabilities. The reproducible research system developed by Schwab et al. (2000[23]) includes not only custom "make" rules but also an obscure and hardly portable agglomeration of shell and Perl scripts that extend "make" (Fomel et al., 1997[24]). Several alternative systems for dependency-checking software construction have been developed in recent years. One of the most promising new tools is SCons, enthusiastically endorsed by Dubois (2003[25]). The SCons initial design won the Software Carpentry competition sponsored by Los Alamos National Laboratory in 2000 in the category of "a dependency management tool to replace make." Some of the main advantages of SCons are:
- SCons configuration files are Python scripts. Python is a modern programming language praised for its readability, elegance, simplicity, and power (Rossum, 2000a[26];Rossum, 2000b[27]). Scales and Ecke (2002[28]) recommend Python as the first programming language for geophysics students.
- SCons offers reliable, automatic, and extensible dependency analysis and creates a global view of all dependencies—no more "make depend," "make clean," or multiple build passes of touching and reordering targets to get all the dependencies.
- SCons has built-in support for many programming languages and systems, including C, C++, Fortran, Java, and LaTeX.
- While "make" relies on timestamps to detect file changes (creating numerous problems on platforms with different system clocks), SCons uses a more reliable detection mechanism, employing MD5 signatures by default. It can detect changes not only in files but also in commands used to build them.
- SCons provides integrated support for parallel builds.
- SCons provides configuration support analogous to the "autoconf" utility for testing the environment on different platforms.
- SCons is designed from the ground up as a cross-platform tool. It works equally well on POSIX systems (Linux, Mac OS X, Solaris, etc.) and Windows.
- The stability of SCons is assured by an incremental development methodology utilizing comprehensive regression tests.
- SCons is publicly released under a liberal open-source license[29].
In this paper, we propose to adopt SCons as a new platform for reproducible research in scientific computing.
Paper organization[edit]
To demonstrate our adoption of SCons for reproducible research, we first describe a couple of simple examples of computational experiments and then show how SCons helps us document our computational results.
Example experiments[edit]
The main SConstruct commands defined in our reproducible research environment are collected in the table.
| Fetch(data_file,dir[,ftp_server_info]) |
| A rule to download data_file from a specific directory dir of an FTP server |
| Flow(target[s],source[s],command[s][,stdin][,stdout]) |
| A rule to generate target[s] from source[s] using command[s] |
| Plot(intermediate_plot[,source],plot_command) or
Plot(intermediate_plot,intermediate_plots,combination) |
| A rule to generate intermediate_plot in the working directory. |
| Result(plot[,source],plot_command) or
Result(plot,intermediate_plots,combination) |
| A rule to generate a final plot in the special Fig folder of the working directory. |
| End() |
| A rule to collect default targets. |
These commands are defined in $PYTHONPATH/rsf/proj.py where RSFROOT is the environmental variable to the Madagascar installation directory. The source of this file is in framework/rsf/proj.py.
Example 1[edit]
To follow the first example, select a working project directory and copy the following code to a file named SConstruct[30].
from rsf.proj import *
# Download the input data file
Fetch('lena.img','imgs')
# Create RSF header
Flow('lena.hdr','lena.img',
'echo n1=512 n2=513 in=$SOURCE data_format=native_uchar',
stdin=0)
# Convert to floating point and window out the first trace
Flow('lena','lena.hdr','dd type=float | window f2=1')
# Display
Result('lena',
'''
sfgrey title="Hello, World!" transp=n color=b bias=128
clip=100 screenratio=1
''')
# Wrap up
End()
This is our "hello world" example that illustrates the basic use of
some of the commands presented in Table~(tbl:commands). The plan
for this experiment is to download data from a public data
server, convert it to an appropriate file format, and generate a
figure for publication. But let us look at the
SConstruct script and try to decorticate it.
from rsf.proj import *
is a standard Python command that loads the Madagascar project
management module rsf/proj.py which provides our extension to
SCons.
Fetch('lena.img','imgs')
instructs SCons to connect to a public data server (the default server
if no FTP server information is provided) and to fetch the data file
lena.img from the data/imgs directory.
Try running "scons lena.img" on the command line. The successful output should look like
bash$ scons lena.img scons: Reading SConscript files ... scons: done reading SConscript files. scons: Building targets ... retrieve(["lena.img"], []) scons: done building targets.
with the target file lena.img appearing in your directory. In the following examples, we will use -Q (quiet) option of scons to suppress the verbose output.
Flow('lena.hdr','lena.img',
'echo n1=512 n2=513 in=$SOURCE data_format=native_uchar',
stdin=0)
prepares the Madagascar header file lena.hdr using the
standard Unix command echo.
bash$ scons -Q lena.hdr echo n1=512 n2=513 in=lena.img data_format=native_uchar > lena.hdr
Since echo does not take a standard input, stdin is set to 0 in the Flow command; otherwise, the first source is the standard input. Likewise, the first target is the standard output unless otherwise specified.
Note that
lena.img is referred as $SOURCE in the command. This
allows us to change the source file's name without changing the command.
The data format of the lena.img image file is uchar
(unsigned character), the image consists of 513 traces with 512
samples per trace. Our next step is to convert the image
representation to floating point numbers and to window out the first
trace so that the final image is 512 by 512 square. The two
transformations are conveniently combined into one with the help of a Unix pipe.
Flow('lena','lena.hdr','dd type=float | window f2=1')
bash$ scons -Q lena scons: *** Do not know how to make target `lena'. Stop.
What happened? In the absence of the file suffix, the Flow command assumes that the target file suffix is ".rsf". Let us try again.
scons -Q lena.rsf < lena.hdr /RSF/bin/sfdd type=float | /RSF/bin/sfwindow f2=1 > lena.rsf
Notice that Madagascar modules sfdd and sfwindow get substituted for the corresponding short names in the SConstruct file. The file lena.rsf is in a regularly sampled format[31] and can be examined, for example, with sfin lena.rsf[32].
bash$ sfin lena.rsf
lena.rsf:
in="/datapath/lena.rsf@"
esize=4 type=float form=native
n1=512 d1=1 o1=0
n2=512 d2=1 o2=1
262144 elements 1048576 bytes
In the last step, we will create a plot file to display the image on the screen and for including it in the publication.
Result('lena',
'''
sfgrey title="Hello, World!" transp=n color=b bias=128
clip=100 screenratio=1
''')
Notice that we broke the long command string into multiple lines by
using Python's triple quote syntax. All the extra white space will be
ignored when the multiple-line string gets translated into the command
line. The Result command has special targets associated with
it. Try, for example, "scons lena.view" to observe the
figure Fig/lena.vpl generated in a specially created
Fig directory and displayed on the screen. The output should
look like this figure.

The reproducible script ends with
End()Ready to experiment? Try some of the following:
- Run scons -c. The -c (clean) option tells SCons to remove all default targets (the Fig/lena.vpl image file in our case) and also all intermediate targets that it generated.
bash$ scons -c -Q Removed lena.img Removed lena.hdr Removed lena.rsf Removed /datapath/lena.rsf@ Removed Fig/lena.vpl
Run scons again, and the default target will be regenerated.
bash$ scons -Q retrieve(["lena.img"], []) echo n1=512 n2=513 in=lena.img data_format=native_uchar > lena.hdr < lena.hdr /RSF/bin/sfdd type=float | /RSF/bin/sfwindow f2=1 > lena.rsf < lena.rsf /RSF/bin/sfgrey title="Hello, World!" transp=n color=b bias=128 clip=100 screenratio=1 > Fig/lena.vpl
- Edit your SConstruct file and change some of the plotting parameters. For example, change the value of clip from clip=100 to clip=50. Run scons again and observe that only the last part of the processing flow (precisely, the part affected by the parameter change) is being run:
bash$ scons -Q view < lena.rsf /RSF/bin/sfgrey title="Hello, World!" transp=n color=b bias=128 clip=50 screenratio=1 > Fig/lena.vpl sfpen Fig/lena.vpl
SCons is smart enough to recognize that your editing did not affect any of the previous results in the data flow chain! Keeping track of dependencies is the main feature that separates data processing and computational experimenting with SCons from using linear shell scripts. This feature can save you a lot of time for computationally demanding data processing and make your experiments more interactive and enjoyable.
- A special parameter to SCons (defined in rsfproj.py) can time the execution of each step in the processing flow. Try running scons TIMER=y.
- The rsfproj module has direct access to the database that stores the parameters of all Madagascar modules. Try running scons CHECKPAR=y to see parameter checking enforced before computations\footnote{This feature is new and experimental and may not work correctly yet}.
The summary of our SCons commands is given in the table.
| scons file |
| Generate file (usually requires .rsf suffix for Flow targets and .vpl suffix for Plot targets.) |
| scons |
| Generate default targets (usually figures specified in Result.) |
| scons view or scons result.view |
| Generate Result figures and display them on the screen. |
| scons print or scons result.print |
| Generate Result figures and print them. |
| scons lock or scons result.lock |
| Generate Result figures and install them in a separate location. |
| scons test or scons result.test |
| Generate Result figures and compare them with the corresponding "locked" figures stored in a separate location (regression testing). |
| scons result.flip |
| Generate the result figure and compare it with the corresponding "locked" figure stored in a separate location by flipping between the two figures on the screen. |
| scons TIMER=y ... |
| Time the execution of each step in the processing flow (using the Unix time utility.) |
| scons CHECKPAR=y ... |
| Check the names and values of all parameters supplied to Madagascar modules in the processing flow before executing anything (guards against incorrect input.) This option is new and experimental. |
Example 2[edit]
The plan for this experiment is to add random noise to the test "Lena" image and then attempt removing it by low-pass filtering and hard thresholding of coefficients in the Fourier domain. The resultant images are shown in the figures.
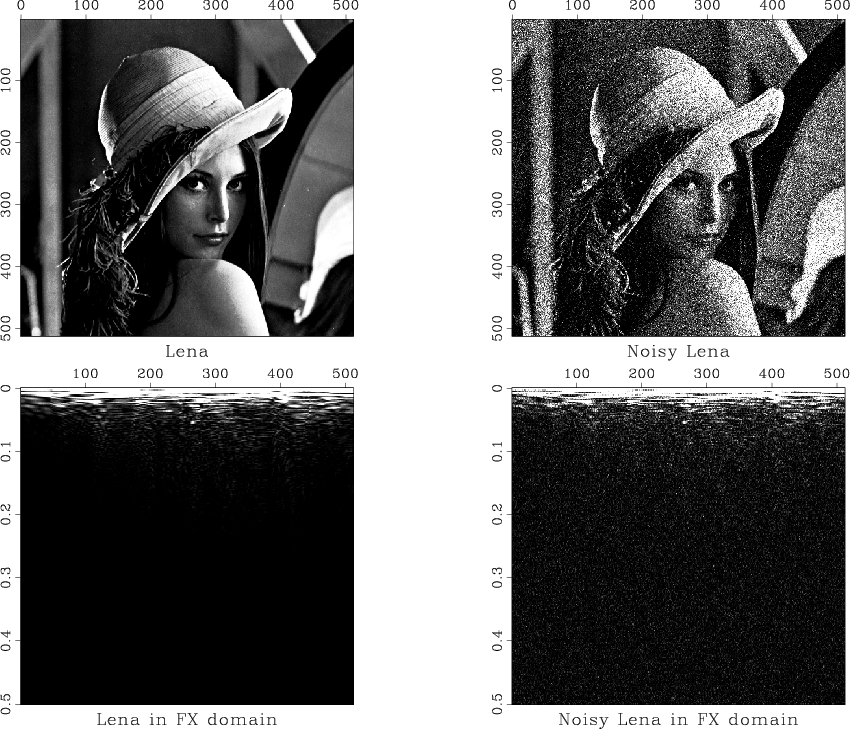
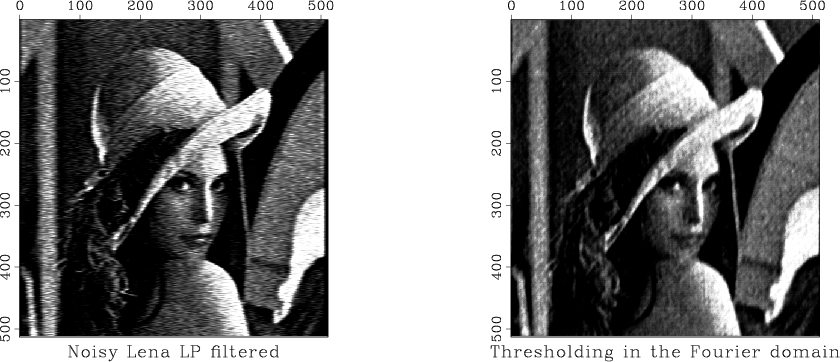
Since the SConstruct| file is a Python script, we can also use all the flexibility and power of the Python language in our Madagascar reproducible scripts. A demo script is available in the rsf/scons/rsfpy subdirectory of the Madagascar book directory. Rather than commenting on it line-by-line, we select some parts of interest. In the SConstruct script, we can declare Python variables
bias = 128and use them later, for example, to define our customized plot command as a Python function
def grey(title,transp='n',bias=bias):
return '''
sfgrey title="%s" transp=%s bias=%g clip=100
screenht=10 screenwd=10 crowd2=0.85 crowd1=0.8
label1= label2=
''' % (title,transp,bias)This Python function, named grey(), can then be called in Plot or Result commands, e.g.
Plot('lplena',grey('Noisy Lena LP filtered'))We can define a Python dictionary, e.g.
titles = {'lena':'Lena',
'nlena':'Noisy Lena'}and loop over its entries, e.g.
for name in titles.keys():
Plot(name,grey(titles[name]) )
cftitle = titles[name]+' in FX domain'
Flow('fx'+name,name,'sfspectra')
Plot('fx'+name,grey(cftitle,'y',100))Note that the title of the plots is obtained by concatenating Python strings. Python strings can also be used to define sequences of commands used in several Flows, e.g.
# 2-D FFT
fft2 = 'sffft1 sym=y | sffft3 sym=y'
Flow('fnlena','nlena',fft2)Finally, in our Madagascar reproducible script, we may want the option to pass command line arguments when running SCons or use default values otherwise, e.g.
# denoising using thresholding in the Fourier domain
fthr = float(ARGUMENTS.get('fthr', 70))
Flow('fthrlena','fnlena','sfthr thr=%f mode="hard"' % fthr)Running scons only, the default value set for fthr (i.e. 70) is used whereas running scons fthr=68 set fthr to a command line specified value. This is by no means an exhaustive list of options, but hopefully, it will give you a flavor of the powerful tool you have in your hands. Enjoy!
Creating reproducible documentation[edit]
You are done with computational experiments and want to communicate them in a paper. SCons helps us create high-quality papers where computational results (figures) are integrated with papers written in LATEX\. The corresponding SCons extension is defined in $PYTHONPATH/rsf/tex.py where RSFROOT is the environmental variable to the Madagascar installation directory. The source of this file is in framework/rsf/tex.py. We summarize the basic methods and commands in the tables.
| Paper(paper_name,[,lclass][,use][,include][,options]) |
| A rule to compile paper_name.tex LATEX\ document using the LATEX2e class specified in lclass (default is geophysics.cls from the SEGTeX package) with additional options specified in options, additional packages specified in use, and additional preamble specified in include. |
| End() |
| A rule to collect default targets (referring to paper.tex document). |
| scons |
| Generate the default target (usually the PDF file paper.pdf from the source LATEX file paper.tex.) |
| scons pdf or scons paper_name.pdf |
| Generate PDF files from LATEX sources paper.tex or paper_name.tex. |
| scons read or scons paper_name.read |
| Generate PDF files from LATEX sources paper.tex or paper_name.tex and display them on the screen. |
| scons print or scons paper_name.print |
| Generate PDF files from LATEX sources paper.tex or paper_name.tex and print them. |
| scons html or scons paper_name.html |
| Generate HTML files from LATEX sources paper.tex or paper_name.tex using LATEXtoHTML. The directory paper_name_html gets created. |
| scons install or scons paper_name.install |
| Generate PDF and HTML files from LATEX sources paper.tex or paper_name.tex and install them in a separate location (used for publishing on a web site). |
| scons wiki or scons paper_name.wiki |
| Convert LATEX sources paper.tex or paper_name.tex to the MediaWiki format (used for publishing on a Wiki web site). |
Example[edit]
This paper by itself is an example of a reproducible document. It is generated using the following SConstruct file which is place in the directory above the projects directories.
from rsf.tex import *
Paper('velan',use='hyperref,listings,color')
End(use='hyperref,listings,color')
This SConstruct generates this paper, but it can also compile
velan.tex in the same directory. Note that there is no
Paper command for paper.tex since it is the default
documentation name. Optional LATEX packages and style used in
paper.tex are passed in the End command.
Let's now take a closer look at paper.tex to understand how the figures of the documentation are linked to the reproducible scripts that created them. First of all, note that paper.tex is not a regular LATEX document but only its body (no documentclass, usepackage, etc.). In our paper, the first figure was created in the project folder easystart (sub-folder of our documentation folder) by the resulting plot lena.vpl. In the LATEX source code, it translates as
\inputdir{easystart}
\sideplot{lena}{height=.25\textheight}{The output of the first numerical experiment.}The inputdir command points to the project directory and the sideplot command calls result_name. The LATEX tag of the figure is fig:result_name. The first time the paper is compiled, the result file is automatically converted to PDF format.
References[edit]
- ↑ Raymond, E. S., 2004, The art of UNIX programming: Addison-Wesley.
- ↑ LeVeque, R. J., to appear, 2006, Wave propagation software, computational science, and reproducible research: Presented at the Proc. International Congress of Mathematicians.
- ↑ Knuth, D. E., 1984, Literate programming: Computer Journal, 27, 97--111.
- ↑ Thimbleby, H., 2003, Explaining code for publication: Software - Practice & Experience, 33, 975--908.
- ↑ http://www.uclic.ucl.ac.uk/harold/
- ↑ Claerbout, J., 1992a, Electronic documents give reproducible research a new meaning: 62nd Ann. Internat. Mtg, 601--604, Soc. of Expl. Geophys.
- ↑ Schwab, M., M. Karrenbach, and J. Claerbout, 2000, Making scientific computations reproducible: Computing in Science & Engineering, 2, 61--67.
- ↑ Buckheit, J. and D. L. Donoho, 1995, Wavelab and reproducible research, in Wavelets and Statistics, volume 103, 55--81. Springer-Verlag.
- ↑ Hubbard, B. B., 1998, The world according to wavelets: The story of a mathematical technique in the making: AK Peters.
- ↑ Mallat, S., 1999, A wavelet tour of signal processing: Academic Press.
- ↑ Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang, 2004, Bioconductor: open software development for computational biology and bioinformatics: Genome Biology, 5, R80.
- ↑ Bivand, R., 2006, Implementing spatial data analysis software tools in r: Geographical Analysis, 38, 23--40.
- ↑ LeVeque, R. J., to appear, 2006, Wave propagation software, computational science, and reproducible research: Presented at the Proc. International Congress of Mathematicians.
- ↑ Stallman, R. M., R. McGrath, and P. D. Smith, 2004, GNU make: A program for directing recompilation: GNU Press.
- ↑ Nichols, D. and S. Cole, 1989, Device independent software installation with CAKE, in SEP-61, 341--344. Stanford Exploration Project.
- ↑ Claerbout, J. F. and D. Nichols, 1990, Why active documents need cake, in SEP-67, 145--148. Stanford Exploration Project.
- ↑ -------- 1992b, How to use Cake with interactive documents, in SEP-73, 451--460. Stanford Exploration Project.
- ↑ Claerbout, J. F. and M. Karrenbach, 1993, How to use cake with interactive documents, in SEP-77, 427--444. Stanford Exploration Project.
- ↑ Schwab, M. and J. Schroeder, 1995, Reproducible research documents using GNUmake, in SEP-89, 217--226. Stanford Exploration Project.
- ↑ Buckheit, J. and D. L. Donoho, 1995, Wavelab and reproducible research, in Wavelets and Statistics, volume 103, 55--81. Springer-Verlag.
- ↑ Sigmon, K. and T. A. Davis, 2001, MATLAB primer, sixth edition: Chapman & Hall.
- ↑ van der Linden, P., 1994, Expert C programming: Prentice Hall.
- ↑ Schwab, M., M. Karrenbach, and J. Claerbout, 2000, Making scientific computations reproducible: Computing in Science & Engineering, 2, 61--67.
- ↑ Fomel, S., M. Schwab, and J. Schroeder, 1997, Empowering SEP's documents, in SEP-94, 339--361. Stanford Exploration Project.
- ↑ Dubois, P. F., 2003, Why Johnny can't build: Computing in Science & Engineering, 5, 83--88.
- ↑ Rossum, G. V., 2000a, Python reference manual: Iuniverse Inc.
- ↑ -------- 2000b, Python tutorial: Iuniverse Inc.
- ↑ Scales, J. A. and H. Ecke, 2002, What programming languages should we teach our undergraduates?: The Leading Edge, 21, 260--267.
- ↑ As of this writing, SCons is in a beta version of 0.96, approaching the 1.0 official release. See http://www.scons.org/.
- ↑ The source of this file is also accessible at $RSFSRC/book/rsf/scons/easystart/SConstruct.
- ↑ See Guide to RSF file format
- ↑ See Guide_to_madagascar_programs#sfin.




