Guide to madagascar API: Difference between revisions
transfer contents |
→Mines JTK: syntax highlighting |
||
| (70 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
<center><font size="-1">''This page was created from the LaTeX source in [http:// | <center><font size="-1">''This page was created from the LaTeX source in [http://sourceforge.net/p/rsf/code/HEAD/tree/trunk/book/rsf/rsf/api.tex book/rsf/rsf/api.tex] using [[latex2wiki]]''</font></center> | ||
This guide explains the RSF programming interface. | [[Image:Fotolia_555071_XS.jpg|right|]] | ||
This guide explains the RSF programming interface. See the '''[[Library_Reference | Library Reference]]''' for more information on how to use the particular APIs. | |||
==Introduction== | ==Introduction== | ||
| Line 9: | Line 11: | ||
It reads and writes RSF files and accesses parameters both from the input file | It reads and writes RSF files and accesses parameters both from the input file | ||
and the command line. The input is processed trace by trace. This is not | and the command line. The input is processed trace by trace. This is not | ||
necessarily the most efficient approach<ref>Compare with the [http://svn.sourceforge.net/ | necessarily the most efficient approach<ref>Compare with the [http://rsf.svn.sourceforge.net/viewvc/rsf/trunk/filt/proc/Mclip.c?view=markup library clip program].</ref> but it suffices for a simple demonstration. | ||
==Installation== | ==Installation== | ||
| Line 19: | Line 21: | ||
scons install | scons install | ||
</pre> | </pre> | ||
The configuration parameters are stored in <tt>$RSFROOT/ | The configuration parameters are stored in <tt>$RSFROOT/share/madagascar/etc/config.py</tt>. | ||
==C interface== | ==C interface== | ||
The C clip function is listed below. | The C clip function is listed below. | ||
<c> | <syntaxhighlight lang="c"> | ||
/* Clip the data. */ | /* Clip the data. */ | ||
| Line 32: | Line 34: | ||
{ | { | ||
int n1, n2, i1, i2; | int n1, n2, i1, i2; | ||
float clip, *trace; | float clip, *trace=NULL; | ||
sf_file in, out; /* Input and output files */ | sf_file in=NULL, out=NULL; /* Input and output files */ | ||
/* Initialize RSF */ | /* Initialize RSF */ | ||
| Line 74: | Line 76: | ||
} | } | ||
free(trace); | |||
sf_close(); | |||
exit(0); | exit(0); | ||
} | } | ||
</ | </syntaxhighlight> | ||
Let us examine it in detail. | Let us examine it in detail. | ||
<c> | <syntaxhighlight lang="c"> | ||
#include <rsf.h> | #include <rsf.h> | ||
</ | </syntaxhighlight> | ||
The include preprocessing directive is required to access the RSF interface. | The include preprocessing directive is required to access the RSF interface. | ||
<c> | <syntaxhighlight lang="c"> | ||
sf_file in, out; /* Input and output files */ | sf_file in=NULL, out=NULL; /* Input and output files */ | ||
</ | </syntaxhighlight> | ||
RSF data files are defined with an abstract <tt>sf_file</tt> data type. An | RSF data files are defined with an abstract <tt>sf_file</tt> data type. An | ||
| Line 94: | Line 98: | ||
<tt>stdio.h</tt> and as close as C gets to an object-oriented style of | <tt>stdio.h</tt> and as close as C gets to an object-oriented style of | ||
programming (Roberts, 1998<ref>Roberts, E. S., 1998, Programming abstractions in C: Addison-Wesley.</ref>). | programming (Roberts, 1998<ref>Roberts, E. S., 1998, Programming abstractions in C: Addison-Wesley.</ref>). | ||
<c> | <syntaxhighlight lang="c"> | ||
/* Initialize RSF */ | /* Initialize RSF */ | ||
sf_init(argc,argv); | sf_init(argc,argv); | ||
</ | </syntaxhighlight> | ||
Before using any of the other functions, you must call | Before using any of the other functions, you must call | ||
<tt>sf_init</tt>. This function parses the command line and | <tt>sf_init</tt>. This function parses the command line and | ||
initializes an internally stored table of command-line parameters. | initializes an internally stored table of command-line parameters. | ||
<c> | <syntaxhighlight lang="c"> | ||
/* standard input */ | /* standard input */ | ||
in = sf_input("in"); | in = sf_input("in"); | ||
/* standard output */ | /* standard output */ | ||
out = sf_output("out"); | out = sf_output("out"); | ||
</ | </syntaxhighlight> | ||
The input and output RSF file objects are created with <tt>sf_input</tt> and | The input and output RSF file objects are created with <tt>sf_input</tt> and | ||
| Line 117: | Line 121: | ||
standard input and <tt>"out"</tt> refers to the file in the standard | standard input and <tt>"out"</tt> refers to the file in the standard | ||
output. | output. | ||
<c> | <syntaxhighlight lang="c"> | ||
/* check that the input is float */ | /* check that the input is float */ | ||
if (SF_FLOAT != sf_gettype(in)) | if (SF_FLOAT != sf_gettype(in)) | ||
sf_error("Need float input"); | sf_error("Need float input"); | ||
</ | </syntaxhighlight> | ||
RSF files can store data of different types (character, integer, | RSF files can store data of different types (character, integer, | ||
| Line 130: | Line 134: | ||
generally a good idea to check the input for user errors and, if they | generally a good idea to check the input for user errors and, if they | ||
cannot be corrected, to take a safe exit. | cannot be corrected, to take a safe exit. | ||
<c> | <syntaxhighlight lang="c"> | ||
/* n1 is the fastest dimension (trace length) */ | /* n1 is the fastest dimension (trace length) */ | ||
if (!sf_histint(in,"n1",&n1)) | if (!sf_histint(in,"n1",&n1)) | ||
| Line 136: | Line 140: | ||
/* leftsize gets n2*n3*n4*... (the number of traces) */ | /* leftsize gets n2*n3*n4*... (the number of traces) */ | ||
n2 = sf_leftsize(in,1); | n2 = sf_leftsize(in,1); | ||
</ | </syntaxhighlight> | ||
Conceptually, the RSF data model is a multidimensional hypercube. By | Conceptually, the RSF data model is a multidimensional hypercube. By | ||
| Line 156: | Line 160: | ||
to extract additional parameters for the hypercube dimensions that we | to extract additional parameters for the hypercube dimensions that we | ||
are not interested in. | are not interested in. | ||
<c> | <syntaxhighlight lang="c"> | ||
/* parameter from the command line (i.e. clip=1.5 ) */ | /* parameter from the command line (i.e. clip=1.5 ) */ | ||
if (!sf_getfloat("clip",&clip)) sf_error("Need clip="); | if (!sf_getfloat("clip",&clip)) sf_error("Need clip="); | ||
</ | </syntaxhighlight> | ||
The clip parameter is read from the command line, where it can be | The clip parameter is read from the command line, where it can be | ||
| Line 167: | Line 171: | ||
found among the command line arguments, the program is aborted with an | found among the command line arguments, the program is aborted with an | ||
error message using the <tt>sf_error</tt> function. | error message using the <tt>sf_error</tt> function. | ||
<c> | <syntaxhighlight lang="c"> | ||
/* allocate floating point array */ | /* allocate floating point array */ | ||
trace = sf_floatalloc (n1); | trace = sf_floatalloc (n1); | ||
</ | </syntaxhighlight> | ||
Next, we allocate an array of floating-point numbers to store a trace | Next, we allocate an array of floating-point numbers to store a trace | ||
| Line 176: | Line 180: | ||
<tt>malloc</tt> the RSF allocation function checks for errors and | <tt>malloc</tt> the RSF allocation function checks for errors and | ||
either terminates the program or returns a valid pointer. | either terminates the program or returns a valid pointer. | ||
<c> | <syntaxhighlight lang="c"> | ||
/* loop over traces */ | /* loop over traces */ | ||
for (i2=0; i2 < n2; i2++) { | for (i2=0; i2 < n2; i2++) { | ||
| Line 192: | Line 196: | ||
sf_floatwrite(trace,n1,out); | sf_floatwrite(trace,n1,out); | ||
} | } | ||
</ | </syntaxhighlight> | ||
The rest of the program is straightforward. We loop over all available | The rest of the program is straightforward. We loop over all available | ||
| Line 201: | Line 205: | ||
specified explicitly in the function name and that the input and | specified explicitly in the function name and that the input and | ||
output files have the RSF type <tt>sf_file</tt>. | output files have the RSF type <tt>sf_file</tt>. | ||
<syntaxhighlight lang="c"> | |||
sf_close(); | |||
exit(0) | |||
</syntaxhighlight> | |||
We explicitly close the input file to avoid leaving a stale temporary file in <tt>$DATAPATH</tt> if the program is called in a pipe sequence. Then, we close the program by sending the shell the Unix code that tells it no errors were encountered. | |||
Note that this is an introductory example, optimized for clarity, not execution speed. For advanced techniques, see [[Madagascar Code Patterns]]. | |||
===Compiling=== | ===Compiling=== | ||
To compile the <tt>clip</tt> program, run | To compile the <tt>clip</tt> program, run | ||
| Line 208: | Line 222: | ||
Change <tt>cc</tt> to the C compiler appropriate for your system and include | Change <tt>cc</tt> to the C compiler appropriate for your system and include | ||
additional compiler flags if necessary. The flags that RSF typically uses are | additional compiler flags if necessary. The flags that RSF typically uses are | ||
in <tt>$RSFROOT/ | in <tt>$RSFROOT/share/madagascar/etc/config.py</tt>. | ||
==C++ interface== | ==C++ interface== | ||
The C++ clip function is listed below. | The C++ clip function is listed below. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
/* Clip the data. */ | /* Clip the data. */ | ||
| Line 249: | Line 263: | ||
exit(0); | exit(0); | ||
} | } | ||
</ | </syntaxhighlight> | ||
Let us examine it line by line. | Let us examine it line by line. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
#include <rsf.hh> | #include <rsf.hh> | ||
</ | </syntaxhighlight> | ||
Including "<tt>rsf.hh</tt>" is required for accessing the RSF C++ | Including "<tt>rsf.hh</tt>" is required for accessing the RSF C++ | ||
interface. | interface. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
sf_init(argc,argv); // Initialize RSF | sf_init(argc,argv); // Initialize RSF | ||
</ | </syntaxhighlight> | ||
A call to <tt>sf_init</tt> is required to initialize the internally stored | A call to <tt>sf_init</tt> is required to initialize the internally stored | ||
table of command-line arguments. | table of command-line arguments. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
iRSF par(0), in; // input parameter, file | iRSF par(0), in; // input parameter, file | ||
oRSF out; // output file | oRSF out; // output file | ||
</ | </syntaxhighlight> | ||
Two classes: <tt>iRSF</tt> and <tt>oRSF</tt> are used to define input and | Two classes: <tt>iRSF</tt> and <tt>oRSF</tt> are used to define input and | ||
output files. For simplicity, the command-line parameters are also handled | output files. For simplicity, the command-line parameters are also handled | ||
as an <tt>iRSF</tt> object, initialized with zero. | as an <tt>iRSF</tt> object, initialized with zero. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
in.get("n1",n1); | in.get("n1",n1); | ||
n2=in.size(1); | n2=in.size(1); | ||
</ | </syntaxhighlight> | ||
Next, we read the data dimensions from the input RSF file object called | Next, we read the data dimensions from the input RSF file object called | ||
| Line 281: | Line 295: | ||
number of traces is the size of <tt>in</tt> remaining after excluding the | number of traces is the size of <tt>in</tt> remaining after excluding the | ||
first dimension. It is extracted with the <tt>size</tt> method. | first dimension. It is extracted with the <tt>size</tt> method. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
par.get("clip",clip); // parameter from the command line | par.get("clip",clip); // parameter from the command line | ||
</ | </syntaxhighlight> | ||
The clip parameter should be specified on the command line, for | The clip parameter should be specified on the command line, for | ||
example, as <tt>clip=10</tt>. It is extracted with the <tt>get</tt> | example, as <tt>clip=10</tt>. It is extracted with the <tt>get</tt> | ||
method of <tt>iRSF</tt> class from the <tt>par</tt> object. | method of <tt>iRSF</tt> class from the <tt>par</tt> object. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
std::valarray<float> trace(n1); | std::valarray<float> trace(n1); | ||
</ | </syntaxhighlight> | ||
The trace object has the single-precision floating-point type and is a | The trace object has the single-precision floating-point type and is a | ||
1-D array of length <tt>n1</tt>. It is declared and allocated using | 1-D array of length <tt>n1</tt>. It is declared and allocated using | ||
the <tt>valarray</tt> template class from the standard C++ library. | the <tt>valarray</tt> template class from the standard C++ library. | ||
<cpp> | <syntaxhighlight lang="cpp"> | ||
for (int i2=0; i2 < n2; i2++) { // loop over traces | for (int i2=0; i2 < n2; i2++) { // loop over traces | ||
in >> trace; // read a trace | in >> trace; // read a trace | ||
| Line 306: | Line 320: | ||
out << trace; // write a trace | out << trace; // write a trace | ||
} | } | ||
</ | </syntaxhighlight> | ||
Next, we loop through the traces, read each trace from <tt>in</tt>, clip it | Next, we loop through the traces, read each trace from <tt>in</tt>, clip it | ||
| Line 317: | Line 331: | ||
Change <tt>c++</tt> to the C++ compiler appropriate for your system and | Change <tt>c++</tt> to the C++ compiler appropriate for your system and | ||
include additional compiler flags if necessary. The flags that RSF typically | include additional compiler flags if necessary. The flags that RSF typically | ||
uses are in <tt>$RSFROOT/ | uses are in <tt>$RSFROOT/share/madagascar/etc/config.py</tt>. | ||
==Fortran-77 interface== | ==Fortran-77 interface== | ||
The Fortran-77 clip function is listed below. | The Fortran-77 clip function is listed below. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
program Clipit | program Clipit | ||
implicit none | implicit none | ||
| Line 363: | Line 377: | ||
stop | stop | ||
end | end | ||
</ | </syntaxhighlight> | ||
Let us examine it in detail. | Let us examine it in detail. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
call sf_init() | call sf_init() | ||
</ | </syntaxhighlight> | ||
The program starts with a call to <tt>sf_init</tt>, which initializes the | The program starts with a call to <tt>sf_init</tt>, which initializes the | ||
command-line interface. | command-line interface. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
in = sf_input("in") | in = sf_input("in") | ||
out = sf_output("out") | out = sf_output("out") | ||
</ | </syntaxhighlight> | ||
The input and output files are created with calls to | The input and output files are created with calls to | ||
| Line 388: | Line 402: | ||
the standard input and <tt>"out"</tt> refers to the file in the | the standard input and <tt>"out"</tt> refers to the file in the | ||
standard output. | standard output. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
if (3 .ne. sf_gettype(in)) | if (3 .ne. sf_gettype(in)) | ||
& call sf_error("Need float input") | & call sf_error("Need float input") | ||
</ | </syntaxhighlight> | ||
RSF files can store data of different types (character, integer, | RSF files can store data of different types (character, integer, | ||
| Line 400: | Line 414: | ||
generally a good idea to check the input for user errors and, if they | generally a good idea to check the input for user errors and, if they | ||
cannot be corrected, to take a safe exit. | cannot be corrected, to take a safe exit. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
if (.not. sf_histint(in,"n1",n1)) then | if (.not. sf_histint(in,"n1",n1)) then | ||
call sf_error("No n1= in input") | call sf_error("No n1= in input") | ||
| Line 407: | Line 421: | ||
end if | end if | ||
n2 = sf_leftsize(in,1) | n2 = sf_leftsize(in,1) | ||
</ | </syntaxhighlight> | ||
Conceptually, the RSF data model is a multidimensional hypercube. By | Conceptually, the RSF data model is a multidimensional hypercube. By | ||
| Line 429: | Line 443: | ||
avoid the need to extract additional parameters for the hypercube | avoid the need to extract additional parameters for the hypercube | ||
dimensions that we are not interested in. | dimensions that we are not interested in. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
if (.not. sf_getfloat("clip",clip)) | if (.not. sf_getfloat("clip",clip)) | ||
& call sf_error("Need clip=") | & call sf_error("Need clip=") | ||
</ | </syntaxhighlight> | ||
The clip parameter is read from the command line, where it can be | The clip parameter is read from the command line, where it can be | ||
| Line 440: | Line 454: | ||
found among the command line arguments, the program is aborted with an | found among the command line arguments, the program is aborted with an | ||
error message using the <tt>sf_error</tt> function. | error message using the <tt>sf_error</tt> function. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
do 10 i2=1, n2 | do 10 i2=1, n2 | ||
call sf_floatread(trace,n1,in) | call sf_floatread(trace,n1,in) | ||
| Line 454: | Line 468: | ||
call sf_floatwrite(trace,n1,out) | call sf_floatwrite(trace,n1,out) | ||
10 continue | 10 continue | ||
</ | </syntaxhighlight> | ||
Finally, we do the actual work: loop over input traces, reading, | Finally, we do the actual work: loop over input traces, reading, | ||
| Line 465: | Line 479: | ||
Change <tt>f77</tt> to the Fortran compiler appropriate for your system and | Change <tt>f77</tt> to the Fortran compiler appropriate for your system and | ||
include additional compiler flags if necessary. The flags that RSF typically | include additional compiler flags if necessary. The flags that RSF typically | ||
uses are in <tt>$RSFROOT/ | uses are in <tt>$RSFROOT/share/madagascar/etc/config.py</tt>. | ||
==Fortran-90 interface== | ==Fortran-90 interface== | ||
The Fortran-90 clip function is listed below. | The Fortran-90 clip function is listed below. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
program Clipit | program Clipit | ||
use rsf | use rsf | ||
| Line 500: | Line 515: | ||
call rsf_write(out,trace) | call rsf_write(out,trace) | ||
end do | end do | ||
deallocate (trace) | |||
end program Clipit | end program Clipit | ||
</ | </syntaxhighlight> | ||
Let us examine it in detail. | Let us examine it in detail. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
use rsf | use rsf | ||
</ | </syntaxhighlight> | ||
The program starts with importing the <tt>rsf</tt> module. | The program starts with importing the <tt>rsf</tt> module. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
call sf_init() ! initialize RSF | call sf_init() ! initialize RSF | ||
</ | </syntaxhighlight> | ||
A call to <tt>sf_init</tt> is needed to initialize the command-line | A call to <tt>sf_init</tt> is needed to initialize the command-line | ||
interface. | interface. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
in = rsf_input() | in = rsf_input() | ||
out = rsf_output() | out = rsf_output() | ||
</ | </syntaxhighlight> | ||
The standard input and output files are initialized with | The standard input and output files are initialized with | ||
| Line 526: | Line 544: | ||
<tt>rsf_input("velocity.rsf")</tt> and <tt>rsf_input("vel")</tt> are | <tt>rsf_input("velocity.rsf")</tt> and <tt>rsf_input("vel")</tt> are | ||
acceptable. | acceptable. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
if (sf_float /= gettype(in)) call sf_error("Need float type") | if (sf_float /= gettype(in)) call sf_error("Need float type") | ||
</ | </syntaxhighlight> | ||
A call to <tt>from_par</tt> extracts the "<tt>n1</tt>" parameter | A call to <tt>from_par</tt> extracts the "<tt>n1</tt>" parameter | ||
| Line 546: | Line 564: | ||
<tt>filesize</tt>, we avoid the need to extract additional parameters | <tt>filesize</tt>, we avoid the need to extract additional parameters | ||
for the hypercube dimensions that we are not interested in. | for the hypercube dimensions that we are not interested in. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
n2 = filesize(in,1) | n2 = filesize(in,1) | ||
</ | </syntaxhighlight> | ||
The clip parameter is read from the command line, where it can be | The clip parameter is read from the command line, where it can be | ||
| Line 554: | Line 572: | ||
value for <tt>clip</tt>, we could specify it with an optional | value for <tt>clip</tt>, we could specify it with an optional | ||
argument, i.e. <tt>call~from_par("clip",clip,default)</tt>. | argument, i.e. <tt>call~from_par("clip",clip,default)</tt>. | ||
<fortran> | <syntaxhighlight lang="fortran"> | ||
allocate (trace (n1)) | allocate (trace (n1)) | ||
| Line 563: | Line 581: | ||
where (trace < -clip) trace = -clip | where (trace < -clip) trace = -clip | ||
</ | </syntaxhighlight> | ||
Finally, we do the actual work: loop over input traces, reading, | Finally, we do the actual work: loop over input traces, reading, | ||
| Line 574: | Line 592: | ||
Change <tt>f90</tt> to the Fortran-90 compiler appropriate for your system and | Change <tt>f90</tt> to the Fortran-90 compiler appropriate for your system and | ||
include additional compiler flags if necessary. The flags that RSF typically | include additional compiler flags if necessary. The flags that RSF typically | ||
uses are in <tt>$RSFROOT/ | uses are in <tt>$RSFROOT/share/madagasacar/etc/config.py</tt>. | ||
The complete specification for the F90 API can be found [[Library_Reference#Fortran_90_API | on the Library Reference page]]. | The complete specification for the F90 API can be found [[Library_Reference#Fortran_90_API | on the Library Reference page]]. | ||
==Python interface== | ==Python interface== | ||
Examples that use the python interface are in the directory $RSFSRC/api/python/test. | |||
The Python script clip.py is:. | |||
<syntaxhighlight lang="python"> | |||
<python> | |||
#!/usr/bin/env python | #!/usr/bin/env python | ||
import numpy | import numpy | ||
import | import m8r | ||
par = | par = m8r.Par() | ||
inp = m8r.Input() | |||
output = | output = m8r.Output() | ||
assert 'float' == | assert 'float' == inp.type | ||
n1 = | n1 = inp.int("n1") | ||
n2 = | n2 = inp.size(1) | ||
assert n1 | assert n1 | ||
| Line 602: | Line 622: | ||
for i2 in xrange(n2): # loop over traces | for i2 in xrange(n2): # loop over traces | ||
inp.read(trace) | |||
trace = numpy.clip(trace,-clip,clip) | trace = numpy.clip(trace,-clip,clip) | ||
output.write(trace) | output.write(trace) | ||
</ | </syntaxhighlight> | ||
Let us examine it in detail. | Let us examine it in detail. | ||
<python> | <syntaxhighlight lang="python"> | ||
import numpy | import numpy | ||
import | import m8r | ||
</ | </syntaxhighlight> | ||
The script starts with importing the <tt>numpy</tt> and | The script starts with importing the <tt>numpy</tt> module and the | ||
<tt> | <tt>m8r</tt> API. | ||
<python> | <syntaxhighlight lang="python"> | ||
par = | par = m8r.Par() | ||
inp = m8r.Input() | |||
output = | output = m8r.Output() | ||
assert 'float' == | assert 'float' == inp.type | ||
</ | </syntaxhighlight> | ||
Next, we initialize the command line interface and the standard input and | Next, we initialize the command line interface and the standard input and | ||
output files. We also make sure that the input file type is floating point. | output files. We also make sure that the input file type is floating point. | ||
<python> | <syntaxhighlight lang="python"> | ||
n1 = input.int("n1") | n1 = input.int("n1") | ||
n2 = input.size(1) | n2 = input.size(1) | ||
assert n1 | assert n1 | ||
</ | </syntaxhighlight> | ||
We extract the "<tt>n1</tt>" parameter from the input file. | We extract the "<tt>n1</tt>" parameter from the input file. | ||
| Line 638: | Line 658: | ||
<tt>n3</tt>, etc. If we are interested in the total number of traces, | <tt>n3</tt>, etc. If we are interested in the total number of traces, | ||
like in the clip example, a shortcut is to use the <tt>size</tt> | like in the clip example, a shortcut is to use the <tt>size</tt> | ||
method of the <tt>Input</tt> | method of the <tt>Input</tt> class. Calling <tt>size(0)</tt> returns | ||
the total number of elements in the hypercube (the product of | the total number of elements in the hypercube (the product of | ||
<tt>n1</tt>, <tt>n2</tt>, etc.), calling <tt>size(1)</tt> returns the | <tt>n1</tt>, <tt>n2</tt>, etc.), calling <tt>size(1)</tt> returns the | ||
| Line 644: | Line 664: | ||
calling <tt>size(2)</tt> returns the product of <tt>n3</tt>, | calling <tt>size(2)</tt> returns the product of <tt>n3</tt>, | ||
<tt>n4</tt>, etc. | <tt>n4</tt>, etc. | ||
<python> | <syntaxhighlight lang="python"> | ||
clip = par.float("clip") | clip = par.float("clip") | ||
assert clip | assert clip | ||
</ | </syntaxhighlight> | ||
The clip parameter is read from the command line, where it can be specified, | The clip parameter is read from the command line, where it can be specified, | ||
for example, as <tt>clip=10</tt>. | for example, as <tt>clip=10</tt>. | ||
<python> | <syntaxhighlight lang="python"> | ||
trace = numpy.zeros(n1,'f') | |||
for i2 in xrange(n2): # loop over traces | for i2 in xrange(n2): # loop over traces | ||
inp.read(trace) | |||
trace = numpy.clip(trace,-clip,clip) | trace = numpy.clip(trace,-clip,clip) | ||
output.write(trace) | output.write(trace) | ||
</python> | </syntaxhighlight> | ||
Finally, we do the actual work: allocate an array to hold a trace and loop | |||
over input traces, reading, clipping, and writing out each trace. | |||
Alternative code use inp.trace to allocate an array and read | |||
the whole file into memory. The loop is no longer needed: | |||
<syntaxhighlight lang="python"> | |||
alltraces=inp.read() | |||
alltraces = numpy.clip(alltraces,-clip,clip) | |||
output.write(alltraces) | |||
</syntaxhighlight> | |||
===Compiling=== | ===Compiling=== | ||
The python script does not require compilation. Simply make sure that | The python script does not require compilation. Simply make sure that | ||
<tt>$RSFROOT/lib</tt> is in <tt>PYTHONPATH</tt> and <tt>LD_LIBRARY_PATH</tt>. | <tt>$RSFROOT/lib</tt> is in <tt>PYTHONPATH</tt> and <tt>LD_LIBRARY_PATH</tt>. | ||
===Using the Python API for interactive development in Jupyter notebooks=== | |||
Jupyter notebooks are a good way to prototype python code, explore data, and to | |||
integrate rich text to describe your code. There are four example notebooks in | |||
$RSFSRC/api/python/test that demonstrate how to use Jupyter notebooks. These are: | |||
{|class="wikitable" | |||
|- | |||
| clip.ipynb || clip.py converted to a Jupyter notebook | |||
|- | |||
|simple_m8r_create_write_filter_read.ipynb || numpy, write rsf, Madagascar commands, read rsf, plot | |||
|- | |||
| tle_edge_preserve.ipynb || reproduces a paper from TLE with numpy and Matplotlib | |||
|- | |||
| file_filter_scons_example.ipynb || advanced m8r options; File, filter, and scons from python | |||
|} | |||
They can be started form the command line with the commands: | |||
<pre> | |||
cd $RSFSRC/api/python/test | |||
jupyter notebook clip.ipynb | |||
</pre> | |||
===Interactive mode usage without graphics=== | ===Interactive mode usage without graphics=== | ||
| Line 669: | Line 720: | ||
<pre> | <pre> | ||
sfmath n1=10 n2= | sfmath n1=10 n2=9 output=x1+x2 > test.rsf | ||
</pre> | </pre> | ||
Then, start the python interpreter and paste the following to its command line: | Then, start the python interpreter and paste the following to its command line: | ||
<python> | <syntaxhighlight lang="python"> | ||
import numpy, | import numpy, m8r | ||
inp = m8r.Input('test.rsf') | |||
n1 = | n1 = inp.int("n1") | ||
n2 = | n2 = inp.int("n2") | ||
data = | data =inp.read(shape=(n2,n1)) | ||
data = data.transpose() # Example of numpy in action | data = data.transpose() # Example of numpy in action | ||
print data | print data | ||
</ | </syntaxhighlight> | ||
You will get | You will get | ||
<pre> | <pre> | ||
[[ | [[ 0. 1. 2. 3. 4. 5. 6. 7. 8.] | ||
[ | [ 1. 2. 3. 4. 5. 6. 7. 8. 9.] | ||
[ | [ 2. 3. 4. 5. 6. 7. 8. 9. 10.] | ||
[ | [ 3. 4. 5. 6. 7. 8. 9. 10. 11.] | ||
[ | [ 4. 5. 6. 7. 8. 9. 10. 11. 12.] | ||
[ | [ 5. 6. 7. 8. 9. 10. 11. 12. 13.] | ||
[ | [ 6. 7. 8. 9. 10. 11. 12. 13. 14.] | ||
[ | [ 7. 8. 9. 10. 11. 12. 13. 14. 15.] | ||
[ | [ 8. 9. 10. 11. 12. 13. 14. 15. 16.] | ||
[ | [ 9. 10. 11. 12. 13. 14. 15. 16. 17.]] | ||
</pre> | </pre> | ||
| Line 708: | Line 758: | ||
Python can plot arrays directly from memory, without having to write a file to disk first. [http://matplotlib.sourceforge.net/ Matplotlib] is one of the [http://en.wikipedia.org/wiki/Category:Free_plotting_software several] packages that accomplish this. To create a figure, execute the code in the previous section, followed by: | Python can plot arrays directly from memory, without having to write a file to disk first. [http://matplotlib.sourceforge.net/ Matplotlib] is one of the [http://en.wikipedia.org/wiki/Category:Free_plotting_software several] packages that accomplish this. To create a figure, execute the code in the previous section, followed by: | ||
<python> | <syntaxhighlight lang="python"> | ||
from pylab import * | from pylab import * | ||
imshow(data) | imshow(data) | ||
| Line 714: | Line 764: | ||
ylabel('Y (m)') | ylabel('Y (m)') | ||
title('Matplotlib example') | title('Matplotlib example') | ||
</ | </syntaxhighlight> | ||
If you want to pop up a figure in an interactive session, after pasting to a Python command line the code shown before, also paste: | If you want to pop up a figure in an interactive session, after pasting to a Python command line the code shown before, also paste: | ||
<python> | <syntaxhighlight lang="python"> | ||
show() | show() | ||
</ | </syntaxhighlight> | ||
You will get Figure 1. The figure will pop up if you run the code in a script too, and the script will stop until the figure is manually closed. You must press the floppy disk button in order to save it. To have the image written to disk automatically, instead of <tt>show()</tt> use: | You will get Figure 1. The figure will pop up if you run the code in a script too, and the script will stop until the figure is manually closed. You must press the floppy disk button in order to save it. To have the image written to disk automatically, instead of <tt>show()</tt> use: | ||
<python> | <syntaxhighlight lang="python"> | ||
savefig('myfile.png') | savefig('myfile.png') | ||
</ | </syntaxhighlight> | ||
[[Image:matplotlib_imshow.png]] | [[Image:matplotlib_imshow.png]] | ||
| Line 732: | Line 782: | ||
Putting it all together, here is a sample script reading a RSF file from stdin and printing out a figure: | Putting it all together, here is a sample script reading a RSF file from stdin and printing out a figure: | ||
<python> | <syntaxhighlight lang="python"> | ||
#!/usr/bin/env python | #!/usr/bin/env python | ||
import | import m8r, numpy, sys, pylab | ||
inp = m8r.Input('test.rsf') | |||
n1 = | n1 = inp.int("n1") | ||
n2 = | n2 = inp.int("n2") | ||
data = | data=input.read() | ||
input.read( | |||
pylab.imshow(data) | pylab.imshow(data) | ||
pylab.savefig('out.png') | pylab.savefig('out.png') | ||
</ | </syntaxhighlight> | ||
===Python interfaces to the standalone programs=== | |||
The <tt>m8r</tt> module offers a way to call Madagascar standalone programs (including graphics) elegantly from inside a Python program, interactively or in batch mode. The blog contains examples of running the <tt>m8r</tt> module [http://www.ahay.org/rsflog/index.php?/archives/173-Extending-Python-interface.html from inside a SAGE notebook] or [http://www.ahay.org/rsflog/index.php?/archives/264-Running-Madagascar-in-an-interactive-console.html from inside an iPython shell]. | |||
==MATLAB interface== | ==MATLAB interface== | ||
The MATLAB clip function is listed below. | The MATLAB clip function is listed below. | ||
<matlab> | <syntaxhighlight lang="matlab"> | ||
function clip(in,out,clip) | function clip(in,out,clip) | ||
%CLIP Clip the data | %CLIP Clip the data | ||
| Line 766: | Line 817: | ||
rsf_write(trace,out,'same'); | rsf_write(trace,out,'same'); | ||
end | end | ||
</syntaxhighlight> | |||
</ | |||
Let us examine it in detail. | Let us examine it in detail. | ||
<matlab> | <syntaxhighlight lang="matlab"> | ||
dims = rsf_dim(in); | dims = rsf_dim(in); | ||
</ | </syntaxhighlight> | ||
We start by figuring out the input file dimensions. | We start by figuring out the input file dimensions. | ||
<matlab> | <syntaxhighlight lang="matlab"> | ||
n1 = dims(1); % trace length | n1 = dims(1); % trace length | ||
n2 = prod(dims(2:end)); % number of traces | n2 = prod(dims(2:end)); % number of traces | ||
</ | </syntaxhighlight> | ||
The first dimension is the trace length, the product of all other | The first dimension is the trace length, the product of all other | ||
dimensions correspond to the number of traces. | dimensions correspond to the number of traces. | ||
<matlab> | <syntaxhighlight lang="matlab"> | ||
trace = 1:n1; % allocate trace | trace = 1:n1; % allocate trace | ||
rsf_create(out,in) % create an output file | rsf_create(out,in) % create an output file | ||
</ | </syntaxhighlight> | ||
Next, we allocate the trace array and create an output file. | Next, we allocate the trace array and create an output file. | ||
<matlab> | <syntaxhighlight lang="matlab"> | ||
for i2 = 1:n2 % loop over traces | for i2 = 1:n2 % loop over traces | ||
rsf_read(trace,in,'same'); | rsf_read(trace,in,'same'); | ||
| Line 795: | Line 845: | ||
rsf_write(trace,out,'same'); | rsf_write(trace,out,'same'); | ||
end | end | ||
</ | </syntaxhighlight> | ||
Finally, we do the actual work: loop over input traces, reading, | Finally, we do the actual work: loop over input traces, reading, | ||
| Line 804: | Line 854: | ||
The MATLAB script does not require compilation. Simply make sure that | The MATLAB script does not require compilation. Simply make sure that | ||
<tt>$RSFROOT/lib</tt> is in <tt>MATLABPATH</tt> and <tt>LD_LIBRARY_PATH</tt>. | <tt>$RSFROOT/lib</tt> is in <tt>MATLABPATH</tt> and <tt>LD_LIBRARY_PATH</tt>. | ||
==Java Interface== | |||
There are two interfaces to Java that are available. New codes should be written to use the SWIG interface ONLY. The older interface (using the MINES JTK) is deprecated, and is only provided while users migrate their code to the new interface. | |||
===SWIG=== | |||
To install the SWIG interface: | |||
1. Download the Java Standard Development Kit (JDK). Installation varies by platform. | |||
2. Create the JAVA_HOME environment variable for your shell. This should point to the directory where the JDK was installed. Example (for Ubuntu 10.04): | |||
<syntaxhighlight lang="bash"> | |||
export JAVA_HOME=/usr/lib/jvm/java-6-opensdk | |||
</syntaxhighlight> | |||
3. Reconfigure Madagascar: | |||
<syntaxhighlight lang="bash"> | |||
./configure API=java | |||
</syntaxhighlight> | |||
4. Reinstall Madagascar (Ignore the compilation warnings for Java files): | |||
<syntaxhighlight lang="bash"> | |||
scons; scons install | |||
</syntaxhighlight> | |||
The installation creates two files: <tt>$RSFROOT/lib/libjrsf.so</tt> and <tt>$RSFROOT/lib/rsf.jar</tt>. | |||
Make sure that you set your LD_LIBRARY_PATH to include <tt>$RSFROOT/lib</tt>. | |||
A short demonstration of the interface follows: | |||
<syntaxhighlight lang="java"> | |||
import rsf.RSF; | |||
import rsf.Input; | |||
import rsf.Output; | |||
/* A simple Java program to clip a dataset. */ | |||
public class Clip { | |||
static { | |||
System.loadLibrary("jrsf"); | |||
} | |||
public static void main(String[] args){ | |||
// Initialize command line argument passing | |||
RSF par = new RSF(args); | |||
// Get the input file name. | |||
Input input = new Input("in"); | |||
Output output = new Output("out"); | |||
// Get the value to clip to. | |||
float clip = par.getFloat("clip",0.0f); | |||
// Read our input header | |||
int n3 = input.getN(3); | |||
int n2 = input.getN(2); | |||
int n1 = input.getN(1); | |||
//Perform clipping operation on a single trace and write out. | |||
float[] data = new float[n1]; | |||
for(int i = 0; i < n3; ++i){ | |||
for(int j = 0; j < n2; ++j){ | |||
input.read(data); | |||
for(int k = 0; k < n1; ++k){ | |||
if (data[k] > clip) data[k] = clip; | |||
else if (data[k] < -clip) data[k] = -clip; | |||
} | |||
output.write(data); | |||
} | |||
} | |||
output.setN(1,n1); | |||
output.setN(2,n2); | |||
output.setN(3,n3); | |||
input.close(); | |||
output.close(); | |||
} | |||
} | |||
</syntaxhighlight> | |||
There are only three classes in the interface: | |||
1.'''RSF''' - The command line argument parser, and the initializer for the native interface. An RSF object MUST be instantiated BEFORE instantiating an Input or Output object. | |||
2. '''Input''' - An object that provides read access to an RSF file. | |||
3. '''Output''' - An object that provides write access to an RSF file. | |||
Additionally, the shared library (libjrsf.so or libjrsf.dll) must be included via the System.loadLibrary("jrsf"); as the first command in the file. | |||
To compile on the command line: | |||
<syntaxhighlight lang="bash"> | |||
javac -cp $RSFROOT/lib Clip.java | |||
</syntaxhighlight> | |||
To include this as part of a SCons script (SConstruct): | |||
<syntaxhighlight lang="python"> | |||
from rsf.proj import * | |||
# Compiles Clip.class | |||
project.Java('.','Clip.java') | |||
Flow('dat',None,'spike n1=1000 n2=100 n3=10 nsp=1 k1=500 l1=1000') | |||
Flow('clipd','dat Clip.class', | |||
''' | |||
%s Clip clip=0.5 | |||
''' % WhereIs('java')) | |||
Flow('test.attr','clipd','sfattr') | |||
</syntaxhighlight> | |||
Note, that we compile <tt>Clip.java</tt> using the SCons Java builder. To execute the command, we have to locate the "java" command, which we do using <tt>WhereIs('java')</tt>. Then we execute the class name, and pass any command line arguments as usual. The files are read from standard in for this program and written to standard out. | |||
Please see the [[Library Reference]] for more details on the functions available in the SWIG API. | |||
'''Note: Additional Java packages can included in SCons automatically by setting the CLASSPATH environment variable to point to the JAR files that they are contained in.''' | |||
===Mines JTK=== | |||
'''THIS INTERFACE IS DEPRECATED AND ONLY PROVIDED AS A REFERENCE. THIS INTERFACE IS NO LONGER SUPPORTED AND WILL BE REMOVED''' | |||
This interface may still be compiled by specifying the MINESJTK environment variable. | |||
The interface to Java is less full featured than others. Presently, it only allows you to read RSF files with fewer than 4-dimensions into Java, and then export RSF files. '''The Java interface does not support reading from standard in, or writing from standard out. Therefore, the Java Interface does not support piping either.''' The Java interface at present treats all values as floats, and does not have support for complex numbers. Java programs are not parsed for self-doc information like other programs are either. Using javadoc may be a viable alternative to creating a parsing engine for Java programs. | |||
Once the build is complete, the JAR file rsf.jar will be added to your $RSFROOT/lib folder. | |||
There are two ways to access the API: | |||
1. From the command line: point the classpath at BOTH compile and runtime to this location on the command line. | |||
<syntaxhighlight lang="bash"> | |||
javac -cp $MINESJTK:$RSFROOT/lib/rsf.jar:. Test.java | |||
java -cp $MINESJTK:$RSFROOT/lib/rsf.jar:. Test arg1=... arg2=... arg3=... | |||
</syntaxhighlight> | |||
2. From within a SConstruct script: BOTH the path for the API ($RSFROOT/lib) and the path to the Mines JTK ($MINESJTK) are automatically added to the environment variable CLASSPATH and JAVACLASSPATH for executions made within an SConstruct. Additionally, any additional classes that are already in the CLASSPATH environmental variable in the shell that you launch from will be added to your classpath. This allows you to include additional libraries automatically within your Java programs. The local directory (.) is also included in the CLASSPATH. | |||
<syntaxhighlight lang="python"> | |||
from rsf.proj import * | |||
# Compiles Clip.class | |||
project.Java('.','Clip.java') | |||
Flow('dat',None,'spike n1=1000 n2=100 n3=10 nsp=1 k1=500') | |||
Flow('clipd','Clip.class dat', | |||
''' | |||
%s ${SOURCES[0].filebase} clip=0.5 | |||
in=${SOURCES[1]} out=$TARGET | |||
''' % WhereIs('java'),stdin=0,stdout=-1) | |||
Flow('test.attr','clipd','sfattr') | |||
End() | |||
</syntaxhighlight> | |||
The interface itself is fairly straightforward. More details on the methods exposed by the API can be found at the [[Library_Reference#Java_API | Library Reference]]. | |||
Data clipping, argument parsing, and IO example: | |||
<syntaxhighlight lang="java"> | |||
import rsf.Par; | |||
import rsf.Reader; | |||
import rsf.Writer; | |||
import rsf.Header; | |||
/* A simple Java program to clip a dataset. | |||
Presently, there is no automatic self-documentation generation for use | |||
with sfdoc. Javadoc may be a better way to generate self-doc for Java | |||
programs. | |||
*/ | |||
public class Clip { | |||
public static void main(String[] args){ | |||
// Initialize command line argument passing | |||
Par par = new Par(args); | |||
// Get the input file name. | |||
String input = par.getString("in",""); | |||
// If the input file name is nothing, then quit! | |||
if (input.equals("")){ | |||
System.out.println("Did not find input file!"); | |||
System.exit(1); | |||
} | |||
//If the output file name is nothing, then quit! | |||
String output = par.getString("out",""); | |||
if (output.equals("")){ | |||
System.out.println("Did not find output file!"); | |||
System.exit(1); | |||
} | |||
// Get the value to clip to. | |||
float clip = par.getFloat("clip",0.0f); | |||
//Read our header file. | |||
Header header = Reader.readHeader(input); | |||
// Read our binary data. | |||
float[][][] data = Reader.readBinary3D(header); | |||
//Initialize our array values. | |||
int n3 = header.getN(3); | |||
int n2 = header.getN(2); | |||
int n1 = header.getN(1); | |||
//Perform clipping operation. | |||
for(int i = 0; i < n3; ++i){ | |||
for(int j = 0; j < n2; ++j){ | |||
for(int k = 0; k < n1; ++k){ | |||
float trace = data[i][j][k]; | |||
if (trace > clip) data[i][j][k] = clip; | |||
else if (trace < -clip) data[i][j][k] = -clip; | |||
} | |||
} | |||
} | |||
//Write our data out, using the same header values to the file | |||
//located at: output. | |||
Writer.writeRSF(header,data,output); | |||
} | |||
} | |||
</syntaxhighlight> | |||
How to read a file: | |||
<syntaxhighlight lang="java"> | |||
import rsf.Header; | |||
import rsf.Reader; | |||
import rsf.Writer; | |||
public class Test { | |||
public static void main(String[] args){ | |||
Header header = Reader.readHeader("junk.rsf"); //To read the header file, just feed in the path relative to the execution directory | |||
System.out.println(header); //The header file will print out if you ask it to, this is good for debugging | |||
float[][] data = Reader.readBinary2D(header); //Now I can manipulate my data | |||
} | |||
} | |||
</syntaxhighlight> | |||
How to write a file: | |||
<syntaxhighlight lang="java"> | |||
import rsf.Header; | |||
import rsf.Reader; | |||
import rsf.Writer; | |||
public class Test { | |||
public static void main(String[] args){ | |||
Header header = Reader.readHeader("test.rsf"); //To read the header file, just feed in the path relative to the execution directory | |||
System.out.println(header); //The header file will print out if you ask it to, this is good for debugging | |||
float[][] data = Reader.readBinary2D(header); //Now I can manipulate my data | |||
//...Do something | |||
Writer.writeRSF(header,data,"test2.rsf"); //Write out my data! | |||
} | |||
} | |||
</syntaxhighlight> | |||
If you want to create a dataset from scratch from within Java, then you can create an array of data, modify the values, create a header, and then write it out to rsf: | |||
<syntaxhighlight lang="java"> | |||
import rsf.Header; | |||
import rsf.Reader; | |||
import rsf.Writer; | |||
public class Test { | |||
public static void main(String[] args){ | |||
int n1 = 20; | |||
int n2 = 40; | |||
float[][] data = new float[n2][n1]; | |||
//The order for dimensions is reversed, because RSF stores them as column-major arrays (see Python API). | |||
//...Do something | |||
Header header = new Header(); | |||
header.setN(1,n1); | |||
/* We set the values using the proper RSF number for the dimension, instead of the Java array index. | |||
Example: RSF Dimension 1, corresponds to array index 0 in Java. | |||
However, we set the values using index 1. The mapping is handled behind the scenes. | |||
*/ | |||
header.setN(2,n2); | |||
header.setDelta(1,0.0); | |||
header.setLabel(1,"time"); | |||
header.setUnits(1,"s"); | |||
Writer.writeRSF(header,data,"junk2.rsf"); //Write out my data! | |||
} | |||
} | |||
</syntaxhighlight> | |||
==References== | ==References== | ||
<references/> | <references/> | ||
Latest revision as of 19:45, 11 October 2021

This guide explains the RSF programming interface. See the Library Reference for more information on how to use the particular APIs.
Introduction[edit]
To work with RSF files in your own programs, you may need to use an appropriate programming interface. We will demonstrate the interface in different languages using a simple example. The example is a clipping program. It reads and writes RSF files and accesses parameters both from the input file and the command line. The input is processed trace by trace. This is not necessarily the most efficient approach[1] but it suffices for a simple demonstration.
Installation[edit]
Only the C interface is installed by default. To install other APIs, use API= parameter in the RSF configuration. For example, to install C++ and Fortran-90 API bindings in addition to the basic package, run
./configure API=c++,fortran-90 scons install
The configuration parameters are stored in $RSFROOT/share/madagascar/etc/config.py.
C interface[edit]
The C clip function is listed below.
/* Clip the data. */
#include <rsf.h>
int main(int argc, char* argv[])
{
int n1, n2, i1, i2;
float clip, *trace=NULL;
sf_file in=NULL, out=NULL; /* Input and output files */
/* Initialize RSF */
sf_init(argc,argv);
/* standard input */
in = sf_input("in");
/* standard output */
out = sf_output("out");
/* check that the input is float */
if (SF_FLOAT != sf_gettype(in))
sf_error("Need float input");
/* n1 is the fastest dimension (trace length) */
if (!sf_histint(in,"n1",&n1))
sf_error("No n1= in input");
/* leftsize gets n2*n3*n4*... (the number of traces) */
n2 = sf_leftsize(in,1);
/* parameter from the command line (i.e. clip=1.5 ) */
if (!sf_getfloat("clip",&clip)) sf_error("Need clip=");
/* allocate floating point array */
trace = sf_floatalloc (n1);
/* loop over traces */
for (i2=0; i2 < n2; i2++) {
/*read a trace */
sf_floatread(trace,n1,in);
/* loop over samples */
for (i1=0; i1 < n1; i1++) {
if (trace[i1] > clip) trace[i1]= clip;
else if (trace[i1] < -clip) trace[i1]=-clip;
}
/* write a trace */
sf_floatwrite(trace,n1,out);
}
free(trace);
sf_close();
exit(0);
}Let us examine it in detail.
#include <rsf.h>The include preprocessing directive is required to access the RSF interface.
sf_file in=NULL, out=NULL; /* Input and output files */RSF data files are defined with an abstract sf_file data type. An abstract data type means that the contents of it are not publicly declared, and all operations on sf_file objects should be performed with library functions. This is analogous to FILE * data type used in stdio.h and as close as C gets to an object-oriented style of programming (Roberts, 1998[2]).
/* Initialize RSF */
sf_init(argc,argv);Before using any of the other functions, you must call sf_init. This function parses the command line and initializes an internally stored table of command-line parameters.
/* standard input */
in = sf_input("in");
/* standard output */
out = sf_output("out");The input and output RSF file objects are created with sf_input and sf_output constructor functions. Both these functions take a string argument. The string may refer to a file name or a file tag. For example, if the command line contains vel=velocity.rsf, then both sf_input("velocity.rsf") and sf_input("vel") are acceptable. Two tags are special: "in" refers to the file in the standard input and "out" refers to the file in the standard output.
/* check that the input is float */
if (SF_FLOAT != sf_gettype(in))
sf_error("Need float input");RSF files can store data of different types (character, integer, floating point, complex). We extract the data type of the input file with the library sf_gettype function and check if it represents floating point numbers. If not, the program is aborted with an error message, using the sf_error function. It is generally a good idea to check the input for user errors and, if they cannot be corrected, to take a safe exit.
/* n1 is the fastest dimension (trace length) */
if (!sf_histint(in,"n1",&n1))
sf_error("No n1= in input");
/* leftsize gets n2*n3*n4*... (the number of traces) */
n2 = sf_leftsize(in,1);Conceptually, the RSF data model is a multidimensional hypercube. By convention, the dimensions of the cube are stored in n1=, n2=, etc. parameters. The n1 parameter refers to the fastest axis. If the input dataset is a collection of traces, n1 refers to the trace length. We extract it using the sf_histint function (integer parameter from history) and abort if no value for n1 is found. We could proceed in a similar fashion, extracting n2, n3, etc. If we are interested in the total number of traces, like in the clip example, a shortcut is to use the sf_leftsize function. Calling sf_leftsize(in,0) returns the total number of elements in the hypercube (the product of n1, n2, etc.), calling sf_leftsize(in,1) returns the number of traces (the product of n2, n3, etc.), calling sf_leftsize(in,2) returns the product of n3, n4, etc. By calling sf_leftsize, we avoid the need to extract additional parameters for the hypercube dimensions that we are not interested in.
/* parameter from the command line (i.e. clip=1.5 ) */
if (!sf_getfloat("clip",&clip)) sf_error("Need clip=");The clip parameter is read from the command line, where it can be specified, for example, as clip=10. The parameter has the float type, therefore we read it with the sf_getfloat function. If no clip= parameter is found among the command line arguments, the program is aborted with an error message using the sf_error function.
/* allocate floating point array */
trace = sf_floatalloc (n1);Next, we allocate an array of floating-point numbers to store a trace with the library sf_floatalloc function. Unlike the standard malloc the RSF allocation function checks for errors and either terminates the program or returns a valid pointer.
/* loop over traces */
for (i2=0; i2 < n2; i2++) {
/*read a trace */
sf_floatread(trace,n1,in);
/* loop over samples */
for (i1=0; i1 < n1; i1++) {
if (trace[i1] > clip) trace[i1]= clip;
else if (trace[i1] < -clip) trace[i1]=-clip;
}
/* write a trace */
sf_floatwrite(trace,n1,out);
}The rest of the program is straightforward. We loop over all available traces, read each trace, clip it and right the output out. The syntax of sf_floatread and sf_floatwrite functions is similar to the syntax of the C standard fread and fwrite function except that the type of the element is specified explicitly in the function name and that the input and output files have the RSF type sf_file.
sf_close();
exit(0)We explicitly close the input file to avoid leaving a stale temporary file in $DATAPATH if the program is called in a pipe sequence. Then, we close the program by sending the shell the Unix code that tells it no errors were encountered.
Note that this is an introductory example, optimized for clarity, not execution speed. For advanced techniques, see Madagascar Code Patterns.
Compiling[edit]
To compile the clip program, run
cc clip.c -I$RSFROOT/include -L$RSFROOT/lib -lrsf -lm
Change cc to the C compiler appropriate for your system and include additional compiler flags if necessary. The flags that RSF typically uses are in $RSFROOT/share/madagascar/etc/config.py.
C++ interface[edit]
The C++ clip function is listed below.
/* Clip the data. */
#include <valarray>
#include <rsf.hh>
int main(int argc, char* argv[])
{
sf_init(argc,argv); // Initialize RSF
iRSF par(0), in; // input parameter, file
oRSF out; // output file
int n1, n2; // trace length, number of traces
float clip;
in.get("n1",n1);
n2=in.size(1);
par.get("clip",clip); // parameter from the command line
std::valarray<float> trace(n1);
for (int i2=0; i2 < n2; i2++) { // loop over traces
in >> trace; // read a trace
for (int i1=0; i1 < n1; i1++) { // loop over samples
if (trace[i1] > clip) trace[i1]=clip;
else if (trace[i1] < -clip) trace[i1]=-clip;
}
out << trace; // write a trace
}
exit(0);
}Let us examine it line by line.
#include <rsf.hh>Including "rsf.hh" is required for accessing the RSF C++ interface.
sf_init(argc,argv); // Initialize RSFA call to sf_init is required to initialize the internally stored table of command-line arguments.
iRSF par(0), in; // input parameter, file
oRSF out; // output fileTwo classes: iRSF and oRSF are used to define input and output files. For simplicity, the command-line parameters are also handled as an iRSF object, initialized with zero.
in.get("n1",n1);
n2=in.size(1);Next, we read the data dimensions from the input RSF file object called in: the trace length is a parameter called "n1" and the number of traces is the size of in remaining after excluding the first dimension. It is extracted with the size method.
par.get("clip",clip); // parameter from the command lineThe clip parameter should be specified on the command line, for example, as clip=10. It is extracted with the get method of iRSF class from the par object.
std::valarray<float> trace(n1);The trace object has the single-precision floating-point type and is a 1-D array of length n1. It is declared and allocated using the valarray template class from the standard C++ library.
for (int i2=0; i2 < n2; i2++) { // loop over traces
in >> trace; // read a trace
for (int i1=0; i1 < n1; i1++) { // loop over samples
if (trace[i1] > clip) trace[i1]=clip;
else if (trace[i1] < -clip) trace[i1]=-clip;
}
out << trace; // write a trace
}Next, we loop through the traces, read each trace from in, clip it and write the output to out.
Compiling[edit]
To compile the C++ program, run
c++ clip.cc -I$RSFROOT/include -L$RSFROOT/lib -lrsf++ -lrsf -lm
Change c++ to the C++ compiler appropriate for your system and include additional compiler flags if necessary. The flags that RSF typically uses are in $RSFROOT/share/madagascar/etc/config.py.
Fortran-77 interface[edit]
The Fortran-77 clip function is listed below.
program Clipit
implicit none
integer n1, n2, i1, i2, in, out
integer sf_input, sf_output, sf_leftsize, sf_gettype
logical sf_getfloat, sf_histint
real clip, trace(1000)
call sf_init()
in = sf_input("in")
out = sf_output("out")
if (3 .ne. sf_gettype(in))
& call sf_error("Need float input")
if (.not. sf_histint(in,"n1",n1)) then
call sf_error("No n1= in input")
else if (n1 > 1000) then
call sf_error("n1 is too long")
end if
n2 = sf_leftsize(in,1)
if (.not. sf_getfloat("clip",clip))
& call sf_error("Need clip=")
do 10 i2=1, n2
call sf_floatread(trace,n1,in)
do 20 i1=1, n1
if (trace(i1) > clip) then
trace(i1)=clip
else if (trace(i1) < -clip) then
trace(i1)=-clip
end if
20 continue
call sf_floatwrite(trace,n1,out)
10 continue
stop
endLet us examine it in detail.
call sf_init()The program starts with a call to sf_init, which initializes the command-line interface.
in = sf_input("in")
out = sf_output("out")The input and output files are created with calls to sf_input and sf_output. Because of the absence of derived types in Fortran-77, we use simple integer pointers to represent RSF files. Both sf_input and sf_output accept a character string, which may refer to a file name or a file tag. For example, if the command line contains vel=velocity.rsf, then both sf_input("velocity.rsf") and sf_input("vel") are acceptable. Two tags are special: "in" refers to the file in the standard input and "out" refers to the file in the standard output.
if (3 .ne. sf_gettype(in))
& call sf_error("Need float input")RSF files can store data of different types (character, integer, floating point, complex). The function sf_gettype checks the type of data stored in the RSF file. We make sure that the type corresponds to floating-point numbers. If not, the program is aborted with an error message, using the sf_error function. It is generally a good idea to check the input for user errors and, if they cannot be corrected, to take a safe exit.
if (.not. sf_histint(in,"n1",n1)) then
call sf_error("No n1= in input")
else if (n1 > 1000) then
call sf_error("n1 is too long")
end if
n2 = sf_leftsize(in,1)Conceptually, the RSF data model is a multidimensional hypercube. By convention, the dimensions of the cube are stored in n1=, n2=, etc. parameters. The n1 parameter refers to the fastest axis. If the input dataset is a collection of traces, n1 refers to the trace length. We extract it using the sf_histint function (integer parameter from history) and abort if no value for n1 is found. Since Fortran-77 cannot easily handle dynamic allocation, we also need to check that n1 is not larger than the size of the statically allocated array. We could proceed in a similar fashion, extracting n2, n3, etc. If we are interested in the total number of traces, like in the clip example, a shortcut is to use the sf_leftsize function. Calling sf_leftsize(in,0) returns the total number of elements in the hypercube (the product of n1, n2, etc.), calling sf_leftsize(in,1) returns the number of traces (the product of n2, n3, etc.), calling sf_leftsize(in,2) returns the product of n3, n4, etc. By calling sf_leftsize, we avoid the need to extract additional parameters for the hypercube dimensions that we are not interested in.
if (.not. sf_getfloat("clip",clip))
& call sf_error("Need clip=")The clip parameter is read from the command line, where it can be specified, for example, as clip=10. The parameter has the float type, therefore we read it with the sf_getfloat function. If no clip= parameter is found among the command line arguments, the program is aborted with an error message using the sf_error function.
do 10 i2=1, n2
call sf_floatread(trace,n1,in)
do 20 i1=1, n1
if (trace(i1) > clip) then
trace(i1)=clip
else if (trace(i1) < -clip) then
trace(i1)=-clip
end if
20 continue
call sf_floatwrite(trace,n1,out)
10 continueFinally, we do the actual work: loop over input traces, reading, clipping, and writing out each trace.
Compiling[edit]
To compile the Fortran-77 program, run
f77 clip.f -L$RSFROOT/lib -lrsff -lrsf -lm
Change f77 to the Fortran compiler appropriate for your system and include additional compiler flags if necessary. The flags that RSF typically uses are in $RSFROOT/share/madagascar/etc/config.py.
Fortran-90 interface[edit]
The Fortran-90 clip function is listed below.
program Clipit
use rsf
implicit none
type (file) :: in, out
integer :: n1, n2, i1, i2
real :: clip
real, dimension (:), allocatable :: trace
call sf_init() ! initialize RSF
in = rsf_input()
out = rsf_output()
if (sf_float /= gettype(in)) call sf_error("Need float type")
call from_par(in,"n1",n1)
n2 = filesize(in,1)
call from_par("clip",clip) ! command-line parameter
allocate (trace (n1))
do i2=1, n2 ! loop over traces
call rsf_read(in,trace)
where (trace > clip) trace = clip
where (trace < -clip) trace = -clip
call rsf_write(out,trace)
end do
deallocate (trace)
end program ClipitLet us examine it in detail.
use rsfThe program starts with importing the rsf module.
call sf_init() ! initialize RSFA call to sf_init is needed to initialize the command-line interface.
in = rsf_input()
out = rsf_output()The standard input and output files are initialized with rsf_input and rsf_output functions. Both functions accept optional arguments. For example, if the command line contains vel=velocity.rsf, then both rsf_input("velocity.rsf") and rsf_input("vel") are acceptable.
if (sf_float /= gettype(in)) call sf_error("Need float type")A call to from_par extracts the "n1" parameter from the input file. Conceptually, the RSF data model is a multidimensional hypercube. The n1 parameter refers to the fastest axis. If the input dataset is a collection of traces, n1 corresponds to the trace length. We could proceed in a similar fashion, extracting n2, n3, etc. If we are interested in the total number of traces, like in the clip example, a shortcut is to use the filesize function. Calling filesize(in) returns the total number of elements in the hypercube (the product of n1, n2, etc.), calling filesize(in,1) returns the number of traces (the product of n2, n3, etc.), calling filesize(in,2) returns the product of n3, n4, etc. By calling filesize, we avoid the need to extract additional parameters for the hypercube dimensions that we are not interested in.
n2 = filesize(in,1)The clip parameter is read from the command line, where it can be specified, for example, as clip=10. If we knew a good default value for clip, we could specify it with an optional argument, i.e. call~from_par("clip",clip,default).
allocate (trace (n1))
do i2=1, n2 ! loop over traces
call rsf_read(in,trace)
where (trace > clip) trace = clip
where (trace < -clip) trace = -clipFinally, we do the actual work: loop over input traces, reading, clipping, and writing out each trace.
Compiling[edit]
To compile the Fortran-90 program, run
f90 clip.f90 -I$RSFROOT/include -L$RSFROOT/lib -lrsff90 -lrsf -lm
Change f90 to the Fortran-90 compiler appropriate for your system and include additional compiler flags if necessary. The flags that RSF typically uses are in $RSFROOT/share/madagasacar/etc/config.py.
The complete specification for the F90 API can be found on the Library Reference page.
Python interface[edit]
Examples that use the python interface are in the directory $RSFSRC/api/python/test.
The Python script clip.py is:.
#!/usr/bin/env python
import numpy
import m8r
par = m8r.Par()
inp = m8r.Input()
output = m8r.Output()
assert 'float' == inp.type
n1 = inp.int("n1")
n2 = inp.size(1)
assert n1
clip = par.float("clip")
assert clip
trace = numpy.zeros(n1,'f')
for i2 in xrange(n2): # loop over traces
inp.read(trace)
trace = numpy.clip(trace,-clip,clip)
output.write(trace)Let us examine it in detail.
import numpy
import m8rThe script starts with importing the numpy module and the m8r API.
par = m8r.Par()
inp = m8r.Input()
output = m8r.Output()
assert 'float' == inp.typeNext, we initialize the command line interface and the standard input and output files. We also make sure that the input file type is floating point.
n1 = input.int("n1")
n2 = input.size(1)
assert n1We extract the "n1" parameter from the input file. Conceptually, the RSF data model is a multidimensional hypercube. The n1 parameter refers to the fastest axis. If the input dataset is a collection of traces, n1 corresponds to the trace length. We could proceed in a similar fashion, extracting n2, n3, etc. If we are interested in the total number of traces, like in the clip example, a shortcut is to use the size method of the Input class. Calling size(0) returns the total number of elements in the hypercube (the product of n1, n2, etc.), calling size(1) returns the number of traces (the product of n2, n3, etc.), calling size(2) returns the product of n3, n4, etc.
clip = par.float("clip")
assert clipThe clip parameter is read from the command line, where it can be specified, for example, as clip=10.
trace = numpy.zeros(n1,'f')
for i2 in xrange(n2): # loop over traces
inp.read(trace)
trace = numpy.clip(trace,-clip,clip)
output.write(trace)Finally, we do the actual work: allocate an array to hold a trace and loop over input traces, reading, clipping, and writing out each trace.
Alternative code use inp.trace to allocate an array and read the whole file into memory. The loop is no longer needed:
alltraces=inp.read()
alltraces = numpy.clip(alltraces,-clip,clip)
output.write(alltraces)
Compiling[edit]
The python script does not require compilation. Simply make sure that $RSFROOT/lib is in PYTHONPATH and LD_LIBRARY_PATH.
Using the Python API for interactive development in Jupyter notebooks[edit]
Jupyter notebooks are a good way to prototype python code, explore data, and to integrate rich text to describe your code. There are four example notebooks in $RSFSRC/api/python/test that demonstrate how to use Jupyter notebooks. These are:
| clip.ipynb | clip.py converted to a Jupyter notebook |
| simple_m8r_create_write_filter_read.ipynb | numpy, write rsf, Madagascar commands, read rsf, plot |
| tle_edge_preserve.ipynb | reproduces a paper from TLE with numpy and Matplotlib |
| file_filter_scons_example.ipynb | advanced m8r options; File, filter, and scons from python |
They can be started form the command line with the commands:
cd $RSFSRC/api/python/test jupyter notebook clip.ipynb
Interactive mode usage without graphics[edit]
Madagascar's Python API can be used interactively too. Create an input dataset with
sfmath n1=10 n2=9 output=x1+x2 > test.rsf
Then, start the python interpreter and paste the following to its command line:
import numpy, m8r
inp = m8r.Input('test.rsf')
n1 = inp.int("n1")
n2 = inp.int("n2")
data =inp.read(shape=(n2,n1))
data = data.transpose() # Example of numpy in action
print dataYou will get
[[ 0. 1. 2. 3. 4. 5. 6. 7. 8.] [ 1. 2. 3. 4. 5. 6. 7. 8. 9.] [ 2. 3. 4. 5. 6. 7. 8. 9. 10.] [ 3. 4. 5. 6. 7. 8. 9. 10. 11.] [ 4. 5. 6. 7. 8. 9. 10. 11. 12.] [ 5. 6. 7. 8. 9. 10. 11. 12. 13.] [ 6. 7. 8. 9. 10. 11. 12. 13. 14.] [ 7. 8. 9. 10. 11. 12. 13. 14. 15.] [ 8. 9. 10. 11. 12. 13. 14. 15. 16.] [ 9. 10. 11. 12. 13. 14. 15. 16. 17.]]
This code will also work in batch mode in a Python script, not only pasted to the interpreter's command line.
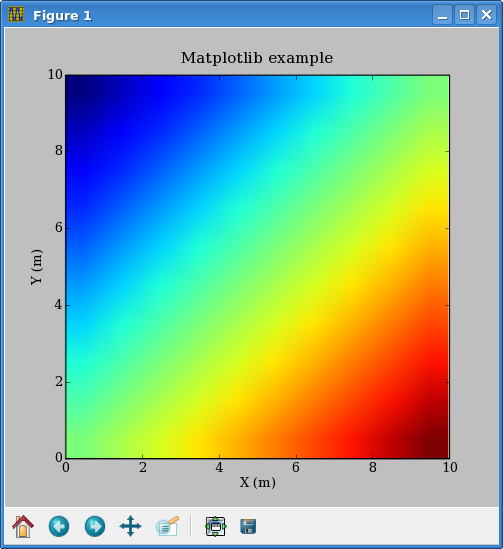
Graphics with Matplotlib[edit]
Python can plot arrays directly from memory, without having to write a file to disk first. Matplotlib is one of the several packages that accomplish this. To create a figure, execute the code in the previous section, followed by:
from pylab import *
imshow(data)
xlabel('X (m)')
ylabel('Y (m)')
title('Matplotlib example')If you want to pop up a figure in an interactive session, after pasting to a Python command line the code shown before, also paste:
show()You will get Figure 1. The figure will pop up if you run the code in a script too, and the script will stop until the figure is manually closed. You must press the floppy disk button in order to save it. To have the image written to disk automatically, instead of show() use:
savefig('myfile.png')Putting it all together, here is a sample script reading a RSF file from stdin and printing out a figure:
#!/usr/bin/env python
import m8r, numpy, sys, pylab
inp = m8r.Input('test.rsf')
n1 = inp.int("n1")
n2 = inp.int("n2")
data=input.read()
pylab.imshow(data)
pylab.savefig('out.png')Python interfaces to the standalone programs[edit]
The m8r module offers a way to call Madagascar standalone programs (including graphics) elegantly from inside a Python program, interactively or in batch mode. The blog contains examples of running the m8r module from inside a SAGE notebook or from inside an iPython shell.
MATLAB interface[edit]
The MATLAB clip function is listed below.
function clip(in,out,clip)
%CLIP Clip the data
dims = rsf_dim(in);
n1 = dims(1); % trace length
n2 = prod(dims(2:end)); % number of traces
trace = 1:n1; % allocate trace
rsf_create(out,in) % create an output file
for i2 = 1:n2 % loop over traces
rsf_read(trace,in,'same');
trace(trace > clip) = clip;
trace(trace < - clip) = -clip;
rsf_write(trace,out,'same');
endLet us examine it in detail.
dims = rsf_dim(in);We start by figuring out the input file dimensions.
n1 = dims(1); % trace length
n2 = prod(dims(2:end)); % number of tracesThe first dimension is the trace length, the product of all other dimensions correspond to the number of traces.
trace = 1:n1; % allocate trace
rsf_create(out,in) % create an output fileNext, we allocate the trace array and create an output file.
for i2 = 1:n2 % loop over traces
rsf_read(trace,in,'same');
trace(trace > clip) = clip;
trace(trace < - clip) = -clip;
rsf_write(trace,out,'same');
endFinally, we do the actual work: loop over input traces, reading, clipping, and writing out each trace.
Available functions[edit]
Only some of the functions in the rsf library have received a MATLAB interface. These functions are rsf_par, rsf_dim, rsf_read, rsf_write and rsf_create. All these functions except rsf_par have been illustrated in the example above.
Compiling[edit]
The MATLAB script does not require compilation. Simply make sure that $RSFROOT/lib is in MATLABPATH and LD_LIBRARY_PATH.
Java Interface[edit]
There are two interfaces to Java that are available. New codes should be written to use the SWIG interface ONLY. The older interface (using the MINES JTK) is deprecated, and is only provided while users migrate their code to the new interface.
SWIG[edit]
To install the SWIG interface:
1. Download the Java Standard Development Kit (JDK). Installation varies by platform.
2. Create the JAVA_HOME environment variable for your shell. This should point to the directory where the JDK was installed. Example (for Ubuntu 10.04):
export JAVA_HOME=/usr/lib/jvm/java-6-opensdk3. Reconfigure Madagascar:
./configure API=java4. Reinstall Madagascar (Ignore the compilation warnings for Java files):
scons; scons installThe installation creates two files: $RSFROOT/lib/libjrsf.so and $RSFROOT/lib/rsf.jar.
Make sure that you set your LD_LIBRARY_PATH to include $RSFROOT/lib.
A short demonstration of the interface follows:
import rsf.RSF;
import rsf.Input;
import rsf.Output;
/* A simple Java program to clip a dataset. */
public class Clip {
static {
System.loadLibrary("jrsf");
}
public static void main(String[] args){
// Initialize command line argument passing
RSF par = new RSF(args);
// Get the input file name.
Input input = new Input("in");
Output output = new Output("out");
// Get the value to clip to.
float clip = par.getFloat("clip",0.0f);
// Read our input header
int n3 = input.getN(3);
int n2 = input.getN(2);
int n1 = input.getN(1);
//Perform clipping operation on a single trace and write out.
float[] data = new float[n1];
for(int i = 0; i < n3; ++i){
for(int j = 0; j < n2; ++j){
input.read(data);
for(int k = 0; k < n1; ++k){
if (data[k] > clip) data[k] = clip;
else if (data[k] < -clip) data[k] = -clip;
}
output.write(data);
}
}
output.setN(1,n1);
output.setN(2,n2);
output.setN(3,n3);
input.close();
output.close();
}
}There are only three classes in the interface:
1.RSF - The command line argument parser, and the initializer for the native interface. An RSF object MUST be instantiated BEFORE instantiating an Input or Output object.
2. Input - An object that provides read access to an RSF file.
3. Output - An object that provides write access to an RSF file.
Additionally, the shared library (libjrsf.so or libjrsf.dll) must be included via the System.loadLibrary("jrsf"); as the first command in the file.
To compile on the command line:
javac -cp $RSFROOT/lib Clip.javaTo include this as part of a SCons script (SConstruct):
from rsf.proj import *
# Compiles Clip.class
project.Java('.','Clip.java')
Flow('dat',None,'spike n1=1000 n2=100 n3=10 nsp=1 k1=500 l1=1000')
Flow('clipd','dat Clip.class',
'''
%s Clip clip=0.5
''' % WhereIs('java'))
Flow('test.attr','clipd','sfattr')Note, that we compile Clip.java using the SCons Java builder. To execute the command, we have to locate the "java" command, which we do using WhereIs('java'). Then we execute the class name, and pass any command line arguments as usual. The files are read from standard in for this program and written to standard out.
Please see the Library Reference for more details on the functions available in the SWIG API.
Note: Additional Java packages can included in SCons automatically by setting the CLASSPATH environment variable to point to the JAR files that they are contained in.
Mines JTK[edit]
THIS INTERFACE IS DEPRECATED AND ONLY PROVIDED AS A REFERENCE. THIS INTERFACE IS NO LONGER SUPPORTED AND WILL BE REMOVED
This interface may still be compiled by specifying the MINESJTK environment variable.
The interface to Java is less full featured than others. Presently, it only allows you to read RSF files with fewer than 4-dimensions into Java, and then export RSF files. The Java interface does not support reading from standard in, or writing from standard out. Therefore, the Java Interface does not support piping either. The Java interface at present treats all values as floats, and does not have support for complex numbers. Java programs are not parsed for self-doc information like other programs are either. Using javadoc may be a viable alternative to creating a parsing engine for Java programs.
Once the build is complete, the JAR file rsf.jar will be added to your $RSFROOT/lib folder.
There are two ways to access the API:
1. From the command line: point the classpath at BOTH compile and runtime to this location on the command line.
javac -cp $MINESJTK:$RSFROOT/lib/rsf.jar:. Test.java
java -cp $MINESJTK:$RSFROOT/lib/rsf.jar:. Test arg1=... arg2=... arg3=...2. From within a SConstruct script: BOTH the path for the API ($RSFROOT/lib) and the path to the Mines JTK ($MINESJTK) are automatically added to the environment variable CLASSPATH and JAVACLASSPATH for executions made within an SConstruct. Additionally, any additional classes that are already in the CLASSPATH environmental variable in the shell that you launch from will be added to your classpath. This allows you to include additional libraries automatically within your Java programs. The local directory (.) is also included in the CLASSPATH.
from rsf.proj import *
# Compiles Clip.class
project.Java('.','Clip.java')
Flow('dat',None,'spike n1=1000 n2=100 n3=10 nsp=1 k1=500')
Flow('clipd','Clip.class dat',
'''
%s ${SOURCES[0].filebase} clip=0.5
in=${SOURCES[1]} out=$TARGET
''' % WhereIs('java'),stdin=0,stdout=-1)
Flow('test.attr','clipd','sfattr')
End()The interface itself is fairly straightforward. More details on the methods exposed by the API can be found at the Library Reference.
Data clipping, argument parsing, and IO example:
import rsf.Par;
import rsf.Reader;
import rsf.Writer;
import rsf.Header;
/* A simple Java program to clip a dataset.
Presently, there is no automatic self-documentation generation for use
with sfdoc. Javadoc may be a better way to generate self-doc for Java
programs.
*/
public class Clip {
public static void main(String[] args){
// Initialize command line argument passing
Par par = new Par(args);
// Get the input file name.
String input = par.getString("in","");
// If the input file name is nothing, then quit!
if (input.equals("")){
System.out.println("Did not find input file!");
System.exit(1);
}
//If the output file name is nothing, then quit!
String output = par.getString("out","");
if (output.equals("")){
System.out.println("Did not find output file!");
System.exit(1);
}
// Get the value to clip to.
float clip = par.getFloat("clip",0.0f);
//Read our header file.
Header header = Reader.readHeader(input);
// Read our binary data.
float[][][] data = Reader.readBinary3D(header);
//Initialize our array values.
int n3 = header.getN(3);
int n2 = header.getN(2);
int n1 = header.getN(1);
//Perform clipping operation.
for(int i = 0; i < n3; ++i){
for(int j = 0; j < n2; ++j){
for(int k = 0; k < n1; ++k){
float trace = data[i][j][k];
if (trace > clip) data[i][j][k] = clip;
else if (trace < -clip) data[i][j][k] = -clip;
}
}
}
//Write our data out, using the same header values to the file
//located at: output.
Writer.writeRSF(header,data,output);
}
}How to read a file:
import rsf.Header;
import rsf.Reader;
import rsf.Writer;
public class Test {
public static void main(String[] args){
Header header = Reader.readHeader("junk.rsf"); //To read the header file, just feed in the path relative to the execution directory
System.out.println(header); //The header file will print out if you ask it to, this is good for debugging
float[][] data = Reader.readBinary2D(header); //Now I can manipulate my data
}
}How to write a file:
import rsf.Header;
import rsf.Reader;
import rsf.Writer;
public class Test {
public static void main(String[] args){
Header header = Reader.readHeader("test.rsf"); //To read the header file, just feed in the path relative to the execution directory
System.out.println(header); //The header file will print out if you ask it to, this is good for debugging
float[][] data = Reader.readBinary2D(header); //Now I can manipulate my data
//...Do something
Writer.writeRSF(header,data,"test2.rsf"); //Write out my data!
}
}If you want to create a dataset from scratch from within Java, then you can create an array of data, modify the values, create a header, and then write it out to rsf:
import rsf.Header;
import rsf.Reader;
import rsf.Writer;
public class Test {
public static void main(String[] args){
int n1 = 20;
int n2 = 40;
float[][] data = new float[n2][n1];
//The order for dimensions is reversed, because RSF stores them as column-major arrays (see Python API).
//...Do something
Header header = new Header();
header.setN(1,n1);
/* We set the values using the proper RSF number for the dimension, instead of the Java array index.
Example: RSF Dimension 1, corresponds to array index 0 in Java.
However, we set the values using index 1. The mapping is handled behind the scenes.
*/
header.setN(2,n2);
header.setDelta(1,0.0);
header.setLabel(1,"time");
header.setUnits(1,"s");
Writer.writeRSF(header,data,"junk2.rsf"); //Write out my data!
}
}References[edit]
- ↑ Compare with the library clip program.
- ↑ Roberts, E. S., 1998, Programming abstractions in C: Addison-Wesley.