Revisiting SEP tour with Madagascar and SCons: Difference between revisions
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<center><font size="-1">''This page was created from the LaTeX source in [http:// | <center><font size="-1">''This page was created from the LaTeX source in [http://sourceforge.net/p/rsf/code/HEAD/tree/trunk/book/rsf/rsf/tour.tex book/rsf/rsf/tour.tex] using [[latex2wiki]]''</font></center> | ||
Many appreciative users were introduced to SEPlib | Many appreciative users were introduced to SEPlib | ||
| Line 25: | Line 25: | ||
Open a file named "SConstruct" in your favorite editor, start it with a | Open a file named "SConstruct" in your favorite editor, start it with a | ||
line | line | ||
<python> | <syntaxhighlight lang="python"> | ||
from rsf.proj import * | from rsf.proj import * | ||
</ | </syntaxhighlight> | ||
and end it with a line | and end it with a line | ||
<python> | <syntaxhighlight lang="python"> | ||
End() | End() | ||
</ | </syntaxhighlight> | ||
The first line tells Python to load the RSF project module. | The first line tells Python to load the RSF project module. | ||
| Line 37: | Line 37: | ||
===Obtaining the test data=== | ===Obtaining the test data=== | ||
Add a Fetch command as follows: | Add a Fetch command as follows: | ||
<python> | <syntaxhighlight lang="python"> | ||
Fetch('Txx.HH','septour') | Fetch('Txx.HH','septour') | ||
</ | </syntaxhighlight> | ||
Now, by running | Now, by running | ||
| Line 47: | Line 47: | ||
you can instruct SCons to connect to an anonymous data server and extract | you can instruct SCons to connect to an anonymous data server and extract | ||
(fetch) the data file "Txx.HH" from the "septour" directory. | (fetch) the data file "Txx.HH" from the "septour" directory. | ||
===Displaying the data=== | ===Displaying the data=== | ||
Add the following line to the <tt>SConstruct</tt> file: | Add the following line to the <tt>SConstruct</tt> file: | ||
<python> | <syntaxhighlight lang="python"> | ||
Result('wiggle0','Txx.HH','wiggle') | Result('wiggle0','Txx.HH','wiggle') | ||
</ | </syntaxhighlight> | ||
| Line 80: | Line 81: | ||
Our next task is to window and plot a significant portion of the data. Add the | Our next task is to window and plot a significant portion of the data. Add the | ||
following line to the <tt>SConstruct</tt> file: | following line to the <tt>SConstruct</tt> file: | ||
<python> | <syntaxhighlight lang="python"> | ||
Flow('windowed','Txx.HH','window n2=10 min1=0.4 max1=0.8') | Flow('windowed','Txx.HH','window n2=10 min1=0.4 max1=0.8') | ||
</ | </syntaxhighlight> | ||
The window command selects the first ten traces and the time window between | The window command selects the first ten traces and the time window between | ||
0.4 and 0.8 seconds. | 0.4 and 0.8 seconds. | ||
We will plot the windowed data with three different plotting programs. | We will plot the windowed data with three different plotting programs. | ||
<python> | <syntaxhighlight lang="python"> | ||
plotpar = ''' | plotpar = ''' | ||
transp=y poly=y yreverse=y pclip=100 nc=100 allpos=n | transp=y poly=y yreverse=y pclip=100 nc=100 allpos=n | ||
| Line 94: | Line 95: | ||
for plot in ('wiggle','contour','grey'): | for plot in ('wiggle','contour','grey'): | ||
Result(plot,'windowed',plot + plotpar) | Result(plot,'windowed',plot + plotpar) | ||
</ | </syntaxhighlight> | ||
For convenience, plotting parameters are put in a string called | For convenience, plotting parameters are put in a string called | ||
| Line 146: | Line 147: | ||
using Fourier interpolation. | using Fourier interpolation. | ||
Subsampling is accomplished with <tt>sfwindow</tt>. | Subsampling is accomplished with <tt>sfwindow</tt>. | ||
<python> | <syntaxhighlight lang="python"> | ||
# decimate time axis by two | # decimate time axis by two | ||
Flow('subsampled','windowed','window j1=2') | Flow('subsampled','windowed','window j1=2') | ||
</ | </syntaxhighlight> | ||
Running <tt>scons -Q subsampled.rsf</tt> produces | Running <tt>scons -Q subsampled.rsf</tt> produces | ||
| Line 167: | Line 168: | ||
#Inverse Fourier transform from frequency to time. | #Inverse Fourier transform from frequency to time. | ||
All three steps are conveniently combined into one using pipes. | All three steps are conveniently combined into one using pipes. | ||
<python> | <syntaxhighlight lang="python"> | ||
# sinc interpolation in the Fourier domain | # sinc interpolation in the Fourier domain | ||
Flow('resampled','subsampled', | Flow('resampled','subsampled', | ||
'fft1 | pad n1=102 | fft1 inv=y opt=n | window max1=0.8') | 'fft1 | pad n1=102 | fft1 inv=y opt=n | window max1=0.8') | ||
</ | </syntaxhighlight> | ||
Why do we pad the Fourier domain to 102? The time length of the original data | Why do we pad the Fourier domain to 102? The time length of the original data | ||
| Line 180: | Line 181: | ||
the figure. Comparing this result with | the figure. Comparing this result with | ||
the previous plots, we can verify a fairly accurate reconstruction. | the previous plots, we can verify a fairly accurate reconstruction. | ||
<python> | <syntaxhighlight lang="python"> | ||
Result('resampled','wiggle title=Resampled' + plotpar) | Result('resampled','wiggle title=Resampled' + plotpar) | ||
</ | </syntaxhighlight> | ||
[[Image:resampled.png|frame|center|To see this figure on your screen, run <tt>scons resampled.view</tt>]] | [[Image:resampled.png|frame|center|To see this figure on your screen, run <tt>scons resampled.view</tt>]] | ||
| Line 193: | Line 194: | ||
windowed data | windowed data | ||
and pipes the result to a wiggle plotting command: | and pipes the result to a wiggle plotting command: | ||
<python> | <syntaxhighlight lang="python"> | ||
Result('nmo','windowed', | Result('nmo','windowed', | ||
''' | ''' | ||
| Line 199: | Line 200: | ||
wiggle pclip=100 max1=0.6 poly=y | wiggle pclip=100 max1=0.6 poly=y | ||
''') | ''') | ||
</ | </syntaxhighlight> | ||
Running <tt>scons -Q nmo.view</tt> produces | Running <tt>scons -Q nmo.view</tt> produces | ||
| Line 213: | Line 214: | ||
point out, the NMO velocity of 2.05~km/s "appears to split the | point out, the NMO velocity of 2.05~km/s "appears to split the | ||
difference between two distinctly non-hyperbolic shear waves". | difference between two distinctly non-hyperbolic shear waves". | ||
[[Image:nmo.png|frame|center|To see this figure on your screen, run <tt>scons | [[Image:nmo.png|frame|center|To see this figure on your screen, run <tt>scons nmo.view</tt>]] | ||
===Advanced plotting=== | ===Advanced plotting=== | ||
| Line 221: | Line 222: | ||
Start by creating common plotting plotting arguments and plotting the | Start by creating common plotting plotting arguments and plotting the | ||
data in greyscale. | data in greyscale. | ||
<python> | <syntaxhighlight lang="python"> | ||
plotpar = plotpar+' min1=.4 max1=.8 max2=1. min2=.05 poly=n' | plotpar = plotpar+' min1=.4 max1=.8 max2=1. min2=.05 poly=n' | ||
Plot('grey','windowed', | Plot('grey','windowed', | ||
'grey wheretitle=t wherexlabel=b' + plotpar) | 'grey wheretitle=t wherexlabel=b' + plotpar) | ||
</ | </syntaxhighlight> | ||
Next, plot the wiggle traces twice: the fist time, using thick black | Next, plot the wiggle traces twice: the fist time, using thick black | ||
lines (<tt>plotcol=0 plotfat=10</tt>), and the second time, using | lines (<tt>plotcol=0 plotfat=10</tt>), and the second time, using | ||
thinner white lines (<tt>plotcol=7 plotfat=5</tt>). | thinner white lines (<tt>plotcol=7 plotfat=5</tt>). | ||
<python> | <syntaxhighlight lang="python"> | ||
Plot('wiggle1','windowed', | Plot('wiggle1','windowed', | ||
'wiggle plotcol=0 plotfat=10' + plotpar) | 'wiggle plotcol=0 plotfat=10' + plotpar) | ||
Plot('wiggle2','windowed', | Plot('wiggle2','windowed', | ||
'wiggle plotcol=7 plotfat=3' + plotpar) | 'wiggle plotcol=7 plotfat=3' + plotpar) | ||
</ | </syntaxhighlight> | ||
The plots are combined by overlaying or by putting them side by side. | The plots are combined by overlaying or by putting them side by side. | ||
<python> | <syntaxhighlight lang="python"> | ||
Result('overplot','grey wiggle1 wiggle2','Overlay') | Result('overplot','grey wiggle1 wiggle2','Overlay') | ||
Result('sidebyside','grey wiggle2','SideBySideIso') | Result('sidebyside','grey wiggle2','SideBySideIso') | ||
</ | </syntaxhighlight> | ||
The resultant plots are shown in the figures. | The resultant plots are shown in the figures. | ||
[[Image:overplot.png|frame|center|To see this figure on your screen, run <tt>scons | [[Image:overplot.png|frame|center|To see this figure on your screen, run <tt>scons overplot.view</tt>]] | ||
[[Image:sidebyside.png|frame|center|To see this figure on your screen, run <tt>scons | [[Image:sidebyside.png|frame|center|To see this figure on your screen, run <tt>scons sidebyside.view</tt>]] | ||
==Conclusions== | ==Conclusions== | ||
| Line 266: | Line 267: | ||
Thanks to Joe Dellinger and Sándor Tálas for creating "SEP | Thanks to Joe Dellinger and Sándor Tálas for creating "SEP | ||
tour" and to James Rickett for updating it. Several generations of | tour" and to James Rickett for updating it. Several generations of | ||
SEP students contributed to SEPlib. We | SEP students contributed to SEPlib. We tried to preserve all their good | ||
ideas when refactoring SEPlib into | ideas when refactoring SEPlib into Madagascar. | ||
The test dataset used in this paper is courtesy of Beltram Nolte and L. Neil Frazer. | The test dataset used in this paper is courtesy of Beltram Nolte and L. Neil Frazer. | ||
| Line 276: | Line 278: | ||
Here is a complete listing of the <tt>SConstruct</tt> file used in this | Here is a complete listing of the <tt>SConstruct</tt> file used in this | ||
example. | example. | ||
<python> | <syntaxhighlight lang="python"> | ||
######################################################### | ######################################################### | ||
# Setting up | # Setting up | ||
| Line 352: | Line 354: | ||
End() | End() | ||
</ | </syntaxhighlight> | ||
Latest revision as of 21:57, 8 April 2014
Many appreciative users were introduced to SEPlib (Claerbout, 1991[1]) by an excellent article of Dellinger and Tálas (1992[2]). In this paper, I show how to create a similar experience using Madagascar and SCons.
Getting started[edit]
Madagascar programs can be piped and executed from the command line, for example:
bash$ sfspike n1=1000 k1=300 title="\s200 Welcome to \c2 RSF" | \ sfbandpass fhi=2 phase=1 | sfwiggle | sfpen
If you are already familiar with SEPlib, you can find most of the familiar programs with the names prepended by "sf". Typing a command without arguments, should produce a concise self-documentation.
bash$ sfbandpass
The recommended way of using RSF, however, is not with the command line but with SCons and "SConstruct" files.
Setting up[edit]
Open a file named "SConstruct" in your favorite editor, start it with a line
from rsf.proj import *and end it with a line
End()The first line tells Python to load the RSF project module.
Obtaining the test data[edit]
Add a Fetch command as follows:
Fetch('Txx.HH','septour')Now, by running
bash$ scons Txx.HH
you can instruct SCons to connect to an anonymous data server and extract (fetch) the data file "Txx.HH" from the "septour" directory.
Displaying the data[edit]
Add the following line to the SConstruct file:
Result('wiggle0','Txx.HH','wiggle')
Note that it does not matter if this line appears before or after the
"Fetch" line. You are simply instructing SCons how to create a
result plot from the input.
Run
bash$ scons wiggle0.view
If everything is setup correctly in your environment, you should see something like the following output in your terminal:
bash$ scons wiggle0.view scons: Reading SConscript files ... scons: done reading SConscript files. scons: Building targets ... retrieve(["Txx.HH"], []) < Txx.HH /path/to/RSF/bin/sfwiggle > Fig/wiggle0.vpl /path/to/RSF/bin/sfpen Fig/wiggle0.vpl
and the following figure appearing on your screen.
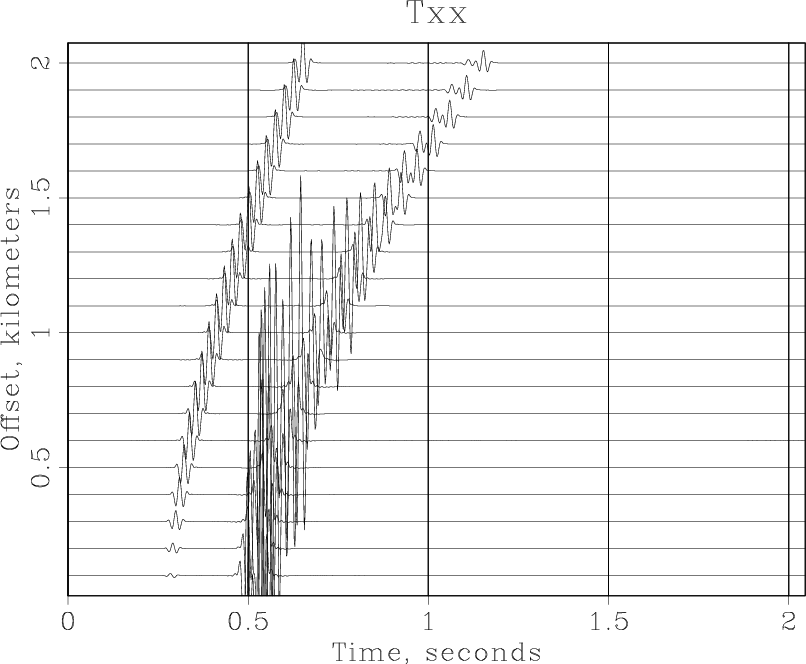
Processing exercises[edit]
Windowing and plotting[edit]
Our next task is to window and plot a significant portion of the data. Add the following line to the SConstruct file:
Flow('windowed','Txx.HH','window n2=10 min1=0.4 max1=0.8')The window command selects the first ten traces and the time window between 0.4 and 0.8 seconds. We will plot the windowed data with three different plotting programs.
plotpar = '''
transp=y poly=y yreverse=y pclip=100 nc=100 allpos=n
'''
for plot in ('wiggle','contour','grey'):
Result(plot,'windowed',plot + plotpar)For convenience, plotting parameters are put in a string called plotpar. A Python string can be enclosed in single, double, or triple quotes. Triple quotes allow the string to span multiple lines. In this case, we use triple quotes for convenience. Next, we loop (using Python's for construct) through three different programs (wiggle, contour, and grey). For each program, the command portion of Result is formed by concatenating two strings with Python's addition operator. Try running scons -Q wiggle.view. You should see something like the following output in your terminal:
bash$ scons -Q wiggle.view < Txx.HH /path/to/RSF/bin/sfwindow n2=10 n1=200 f1=200 > windowed.rsf < windowed.rsf /path/to/RSF/bin/sfwiggle transp=y poly=y yreverse=y pclip=100 nc=200 > Fig/wiggle.vpl /path/to/RSF/bin/sfpen Fig/wiggle.vpl
and a figure similar to Figure~(fig:wiggle) appearing on your screen. The -Q switch tells SCons to run in a quiet mode, suppressing verbose comments. We will use it from now on to save space. You can dismiss the figure by using the "q" key on the keyboard or by hitting the "quit" button. Run scons -Q view, and you should see simply
bash$ scons -Q view /path/to/RSF/bin/sfpen Fig/wiggle.vpl
Since the wiggle.vpl figure is up to date, SCons does not rebuild it. After quitting the figure, SCons will resume processing with
< windowed.rsf /path/to/RSF/bin/sfcontour transp=y poly=y yreverse=y pclip=100 nc=200 > Fig/contour.vpl /path/to/RSF/bin/sfpen Fig/contour.vpl
and a figure appearing on your screen. Quitting the figure, produces
< windowed.rsf /path/to/RSF/bin/sfgrey transp=y poly=y yreverse=y pclip=100 nc=200 > Fig/grey.vpl /path/to/RSF/bin/sfpen Fig/grey.vpl
and the next figure.

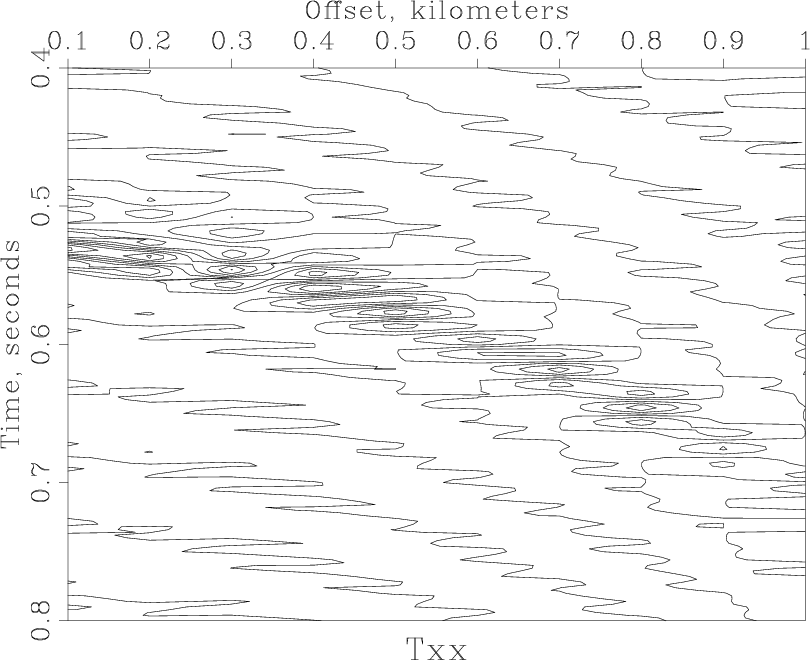
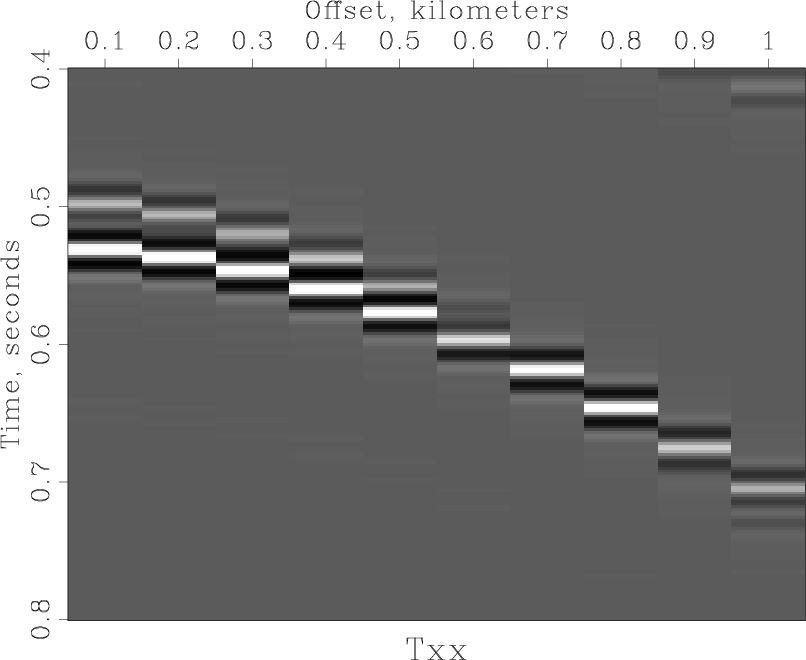
Resampling[edit]
The next example demonstrated simple signal processing using the Fast Fourier Transform. We will first subsample the original data and then recover the data using Fourier interpolation. Subsampling is accomplished with sfwindow.
# decimate time axis by two
Flow('subsampled','windowed','window j1=2')Running scons -Q subsampled.rsf produces
< windowed.rsf /path/to/RSF/bin/sfwindow j1=2 > subsampled.rsf
We can verify that the size of the first axis has decreased by running
sfin windowed.rsf subsampled.rsf.
Try also sfwiggle < subsampled.rsf | sfpen to quickly inspect the subsampled data on the screen. To interpolate the data back to the original sampling, the following sequence of steps can be applied:
- Fourier transform from time domain to frequency domain.
- Pad the frequency axis
- Inverse Fourier transform from frequency to time.
All three steps are conveniently combined into one using pipes.
# sinc interpolation in the Fourier domain
Flow('resampled','subsampled',
'fft1 | pad n1=102 | fft1 inv=y opt=n | window max1=0.8')Why do we pad the Fourier domain to 102? The time length of the original data is 201 samples. In the frequency domain, it can be represented with 101 positive frequencies plus the zero frequency, which amounts to 102. Note that the output of sffft1 does not contain negative frequencies. Finally, we display the result. The reconstructed data is shown in the figure. Comparing this result with the previous plots, we can verify a fairly accurate reconstruction.
Result('resampled','wiggle title=Resampled' + plotpar)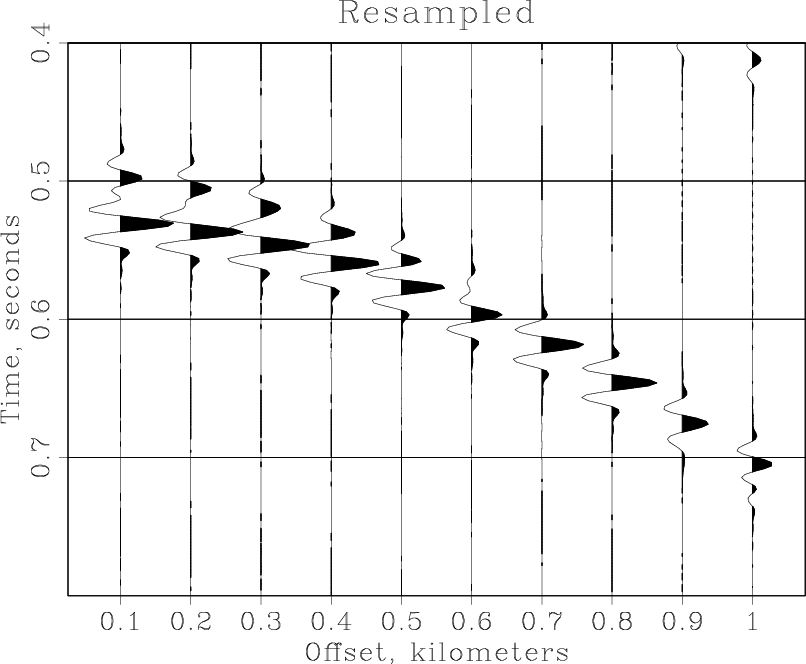
As an exercise, try subsampling the data by a factor of 4 and see if you can still reconstruct the original data with the Fourier method.
Normal Moveout[edit]
The next example applies a simple constant-velocity NMO correction to the windowed data and pipes the result to a wiggle plotting command:
Result('nmo','windowed',
'''
nmostretch v0=2.05 half=n |
wiggle pclip=100 max1=0.6 poly=y
''')Running scons -Q nmo.view produces
< windowed.rsf /path/to/RSF/bin/sfnmostretch v0=2.05 half=n | /path/to/RSF/bin/sfwiggle pclip=100 max1=0.6 poly=y > Fig/nmo.vpl /path/to/RSF/bin/sfpen Fig/nmo.vpl
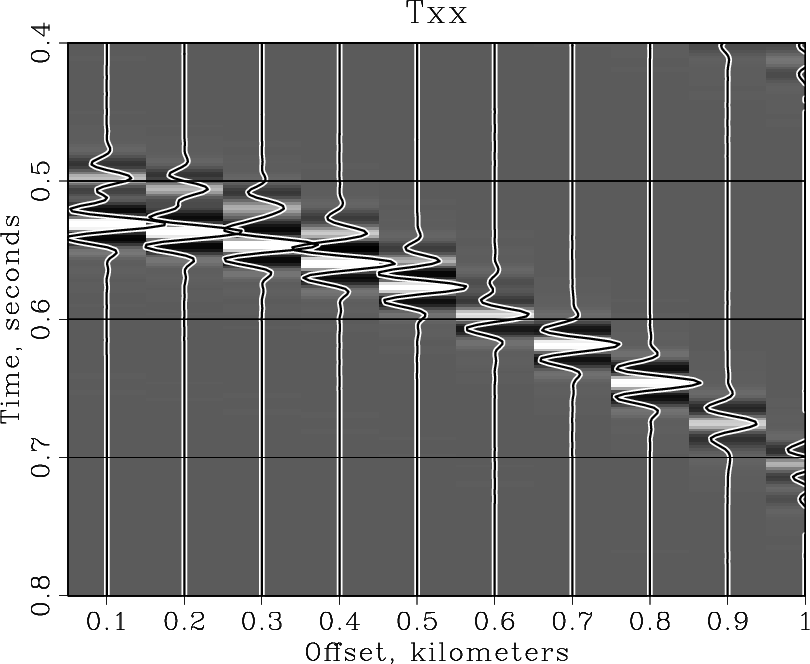
Note that SCons does not recreate the windowed.rsf file if that file is up to date. You can experiment with the NMO velocity (2.05~km/s) or with plotting parameters to get different results. As Dellinger and Tálas (1992[3]) point out, the NMO velocity of 2.05~km/s "appears to split the difference between two distinctly non-hyperbolic shear waves".
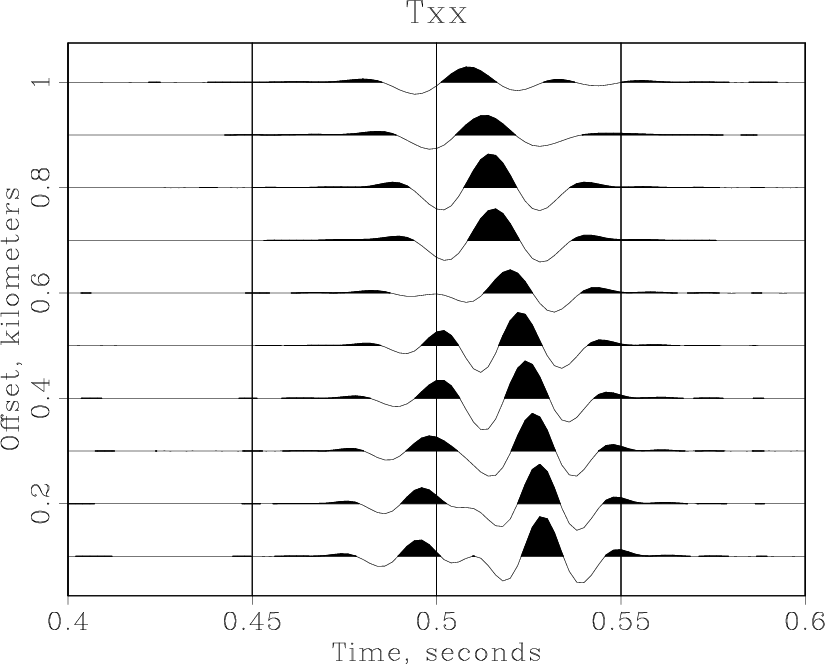
Advanced plotting[edit]
Sometimes, we need to combine different plots either by overlaying them on top of each other or by putting them side by side. Here is an example of accomplishing it with RSF and SCons. Start by creating common plotting plotting arguments and plotting the data in greyscale.
plotpar = plotpar+' min1=.4 max1=.8 max2=1. min2=.05 poly=n'
Plot('grey','windowed',
'grey wheretitle=t wherexlabel=b' + plotpar)Next, plot the wiggle traces twice: the fist time, using thick black lines (plotcol=0 plotfat=10), and the second time, using thinner white lines (plotcol=7 plotfat=5).
Plot('wiggle1','windowed',
'wiggle plotcol=0 plotfat=10' + plotpar)
Plot('wiggle2','windowed',
'wiggle plotcol=7 plotfat=3' + plotpar)The plots are combined by overlaying or by putting them side by side.
Result('overplot','grey wiggle1 wiggle2','Overlay')
Result('sidebyside','grey wiggle2','SideBySideIso')The resultant plots are shown in the figures.
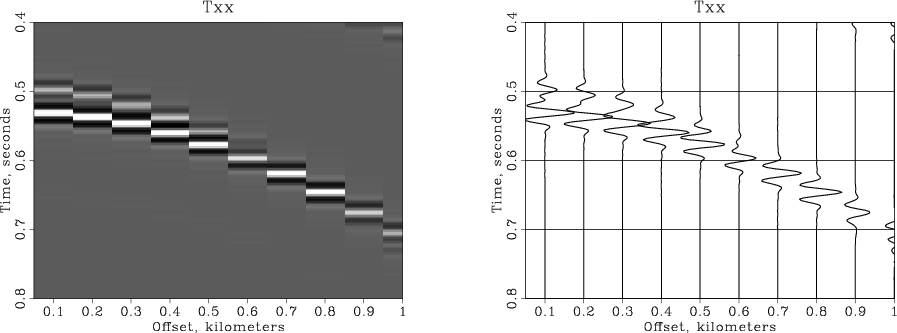
Conclusions[edit]
This tour is not designed as a comprehensive manual. It simply gives a glimpse into working in a reproducible research environment with Madagascar and SCons. The reader is encouraged to experiment with the SConstruct file attached to this tour and included in the Appendix. For other documentation on Madagascar, please see
- Introduction to madagascar
- Installation instructions
- Self-documentation reference for RSF programs
- Guide to madagascar programs
- Guide to RSF file format
- guide to Madagascar programming interface
- Guide to programming with madagascar
Acknowledgments[edit]
Thanks to Joe Dellinger and Sándor Tálas for creating "SEP tour" and to James Rickett for updating it. Several generations of SEP students contributed to SEPlib. We tried to preserve all their good ideas when refactoring SEPlib into Madagascar.
The test dataset used in this paper is courtesy of Beltram Nolte and L. Neil Frazer.
References[edit]
- ↑ Claerbout, J. F., 1991, Introduction to Seplib and SEP utility software, in SEP-70, 413--436. Stanford Exploration Project.
- ↑ Dellinger, J. and S. Tálas, 1992, A tour of SEPlib for new users, in SEP-73, 461--502. Stanford Exploration Project.
- ↑ Dellinger, J. and S. Tálas, 1992, A tour of SEPlib for new users, in SEP-73, 461--502. Stanford Exploration Project.
SConstruct file[edit]
Here is a complete listing of the SConstruct file used in this example.
#########################################################
# Setting up
#########################################################
from rsf.proj import *
#########################################################
# Obtaining the test data
#########################################################
Fetch('Txx.HH','septour')
#########################################################
# Displaying the data
#########################################################
Result('wiggle0','Txx.HH','wiggle')
#########################################################
# Windowing and plotting
#########################################################
Flow('windowed','Txx.HH','window n2=10 min1=0.4 max1=0.8')
plotpar = '''
transp=y poly=y yreverse=y pclip=100 nc=100 allpos=n
'''
for plot in ('wiggle','contour','grey'):
Result(plot,'windowed',plot + plotpar)
#########################################################
# Resampling
#########################################################
# decimate time axis by two
Flow('subsampled','windowed','window j1=2')
# sinc interpolation in the Fourier domain
Flow('resampled','subsampled',
'fft1 | pad n1=102 | fft1 inv=y opt=n | window max1=0.8')
Result('resampled','wiggle title=Resampled' + plotpar)
#########################################################
# Velocity analysis and NMO
#########################################################
Result('nmo','windowed',
'''
nmostretch v0=2.05 half=n |
wiggle pclip=100 max1=0.6 poly=y
''')
#########################################################
# Advanced plotting
#########################################################
plotpar = plotpar+' min1=.4 max1=.8 max2=1. min2=.05 poly=n'
Plot('grey','windowed',
'grey wheretitle=t wherexlabel=b' + plotpar)
Plot('wiggle1','windowed',
'wiggle plotcol=0 plotfat=10' + plotpar)
Plot('wiggle2','windowed',
'wiggle plotcol=7 plotfat=3' + plotpar)
Result('overplot','grey wiggle1 wiggle2','Overlay')
Result('sidebyside','grey wiggle2','SideBySideIso')
#########################################################
# Wrapping up
#########################################################
End()