Several enhancements have been added to Madagascar’s Python interface.
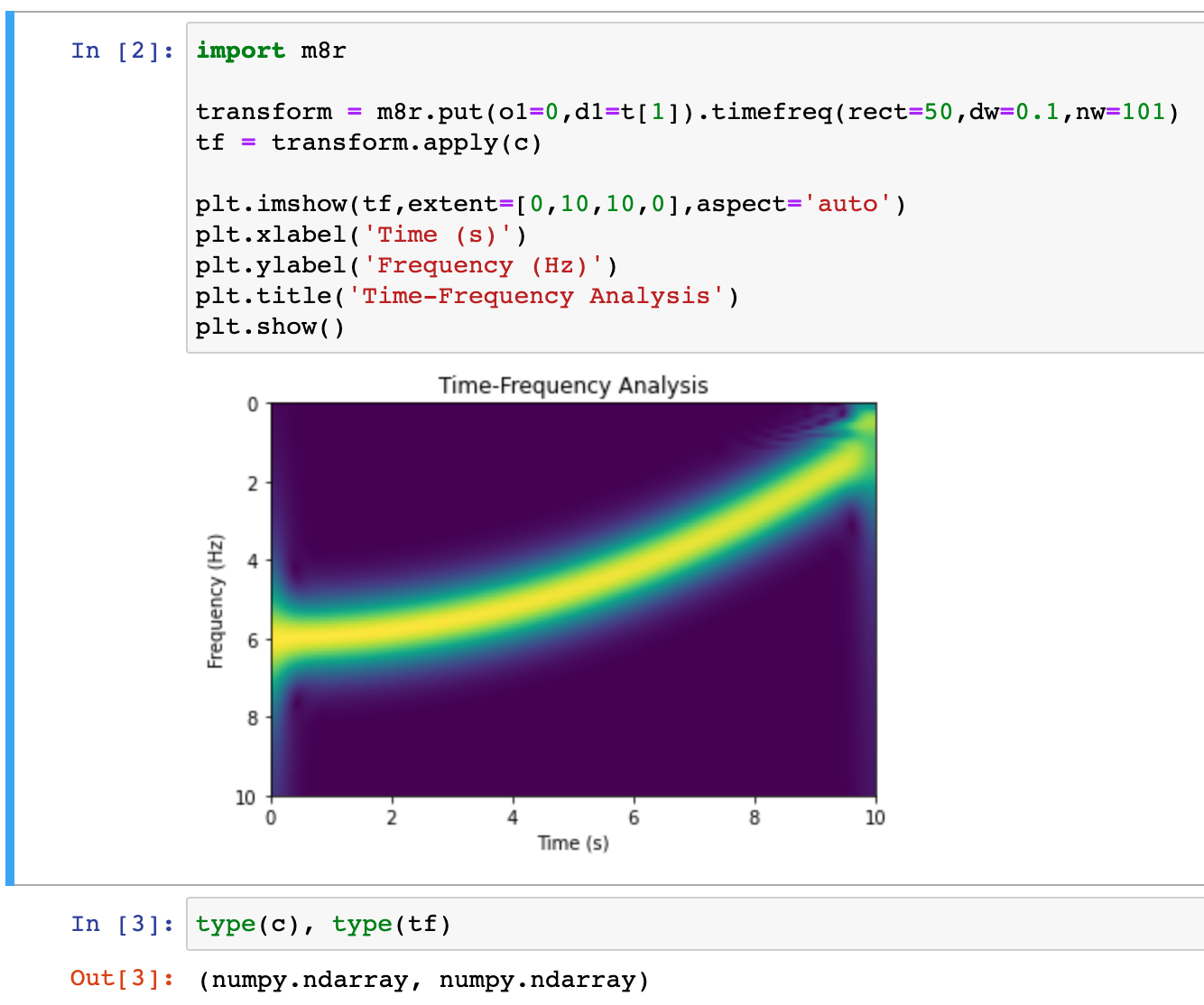
Behind the scene, temporary files are created, and Madgascar programs run in the usual way, but, for the user, they appears like native Python functions. This way, the full power of Madagascar becomes available to people who prefer to work on data analysis projects in a Python environment.
- However, there is no good reason to abandon Madagascar’s use of SCons for managing data analysis workflows even when working in a Python framework. Because
SConstruct scripts are written in Python, they are easy to adapt for including Python functions in place of command-line instructions. See an example of using Keras with SCons or an example of using PyTorch with SCons.
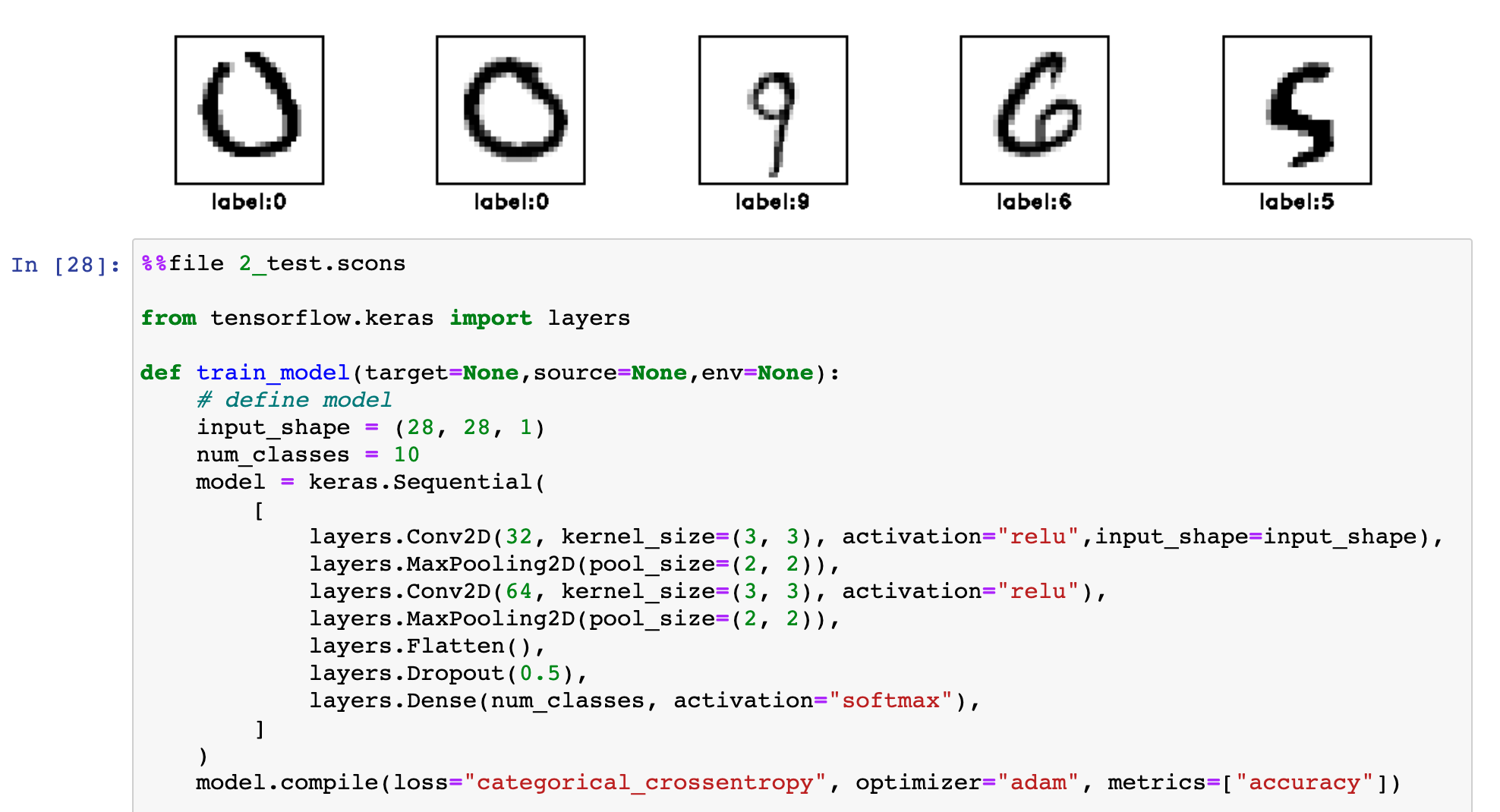
In deep learning projects, the training data, the neural-network model, and the testing data can be treated as files and handled effectively through SCons workflows while mixing with Madagascar commands and workflows.
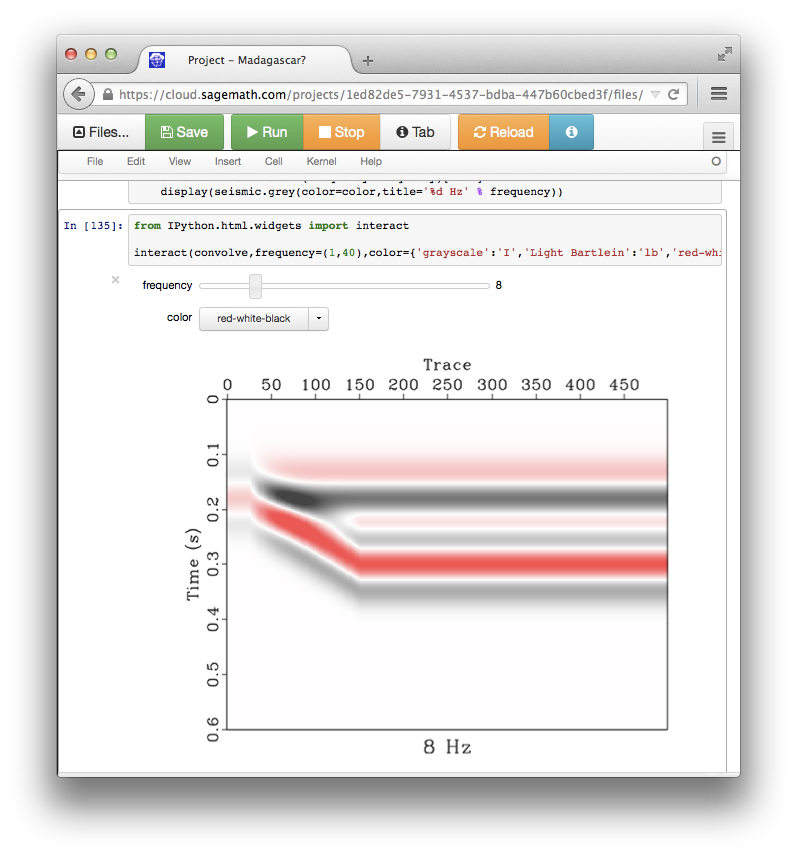
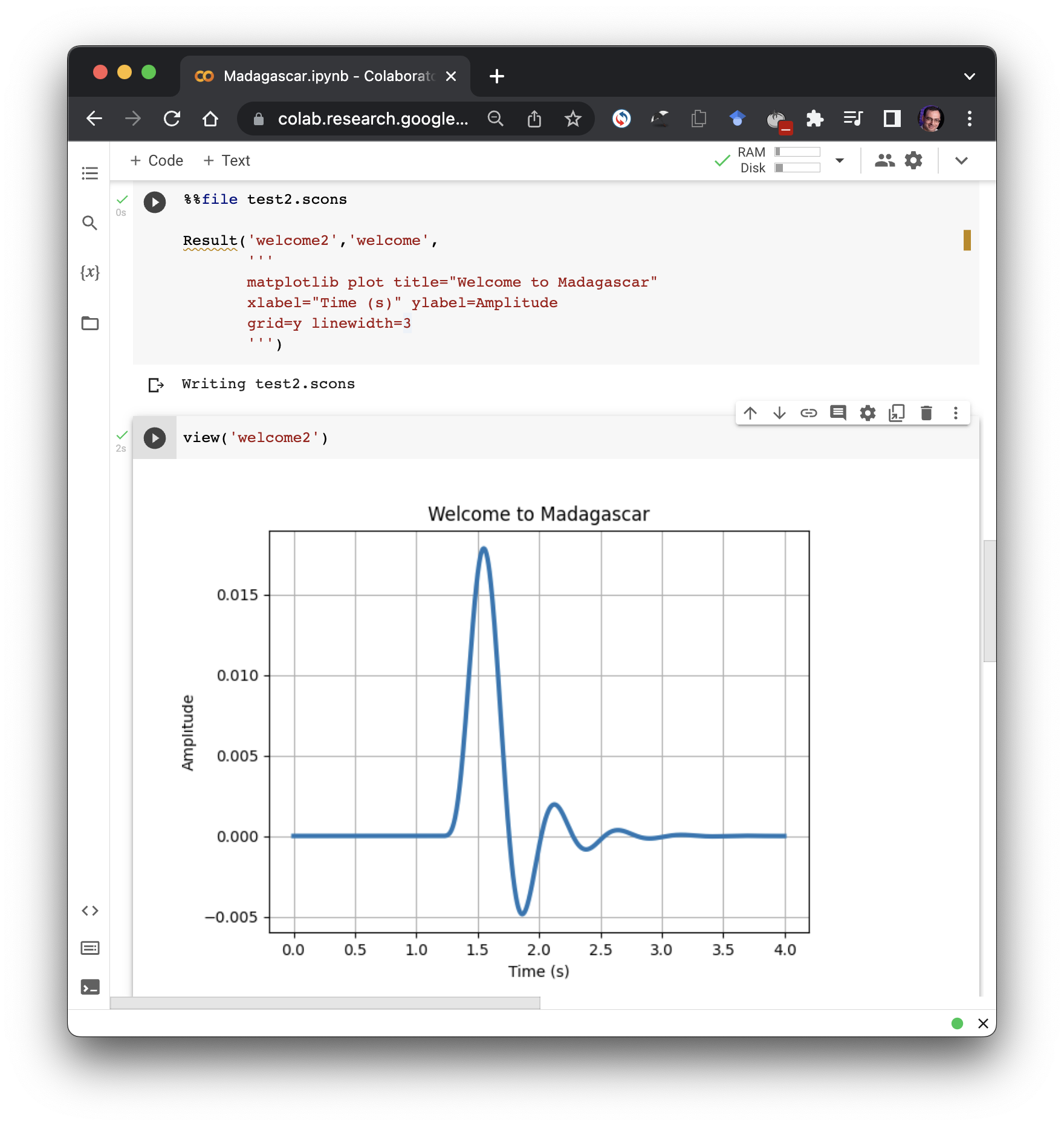
- Plotting with Matplotlib may offer some advanced functionality in comparison with Vplot, such as the possibility of using $\LaTeX$ code in figure labels. It is now possible to use Matplotlib plots in papers reproducible with Madagascar through an application of sfmatplotlib. The figures will be saved in the PDF format and included in reproducible papers in the usual way. See an example.
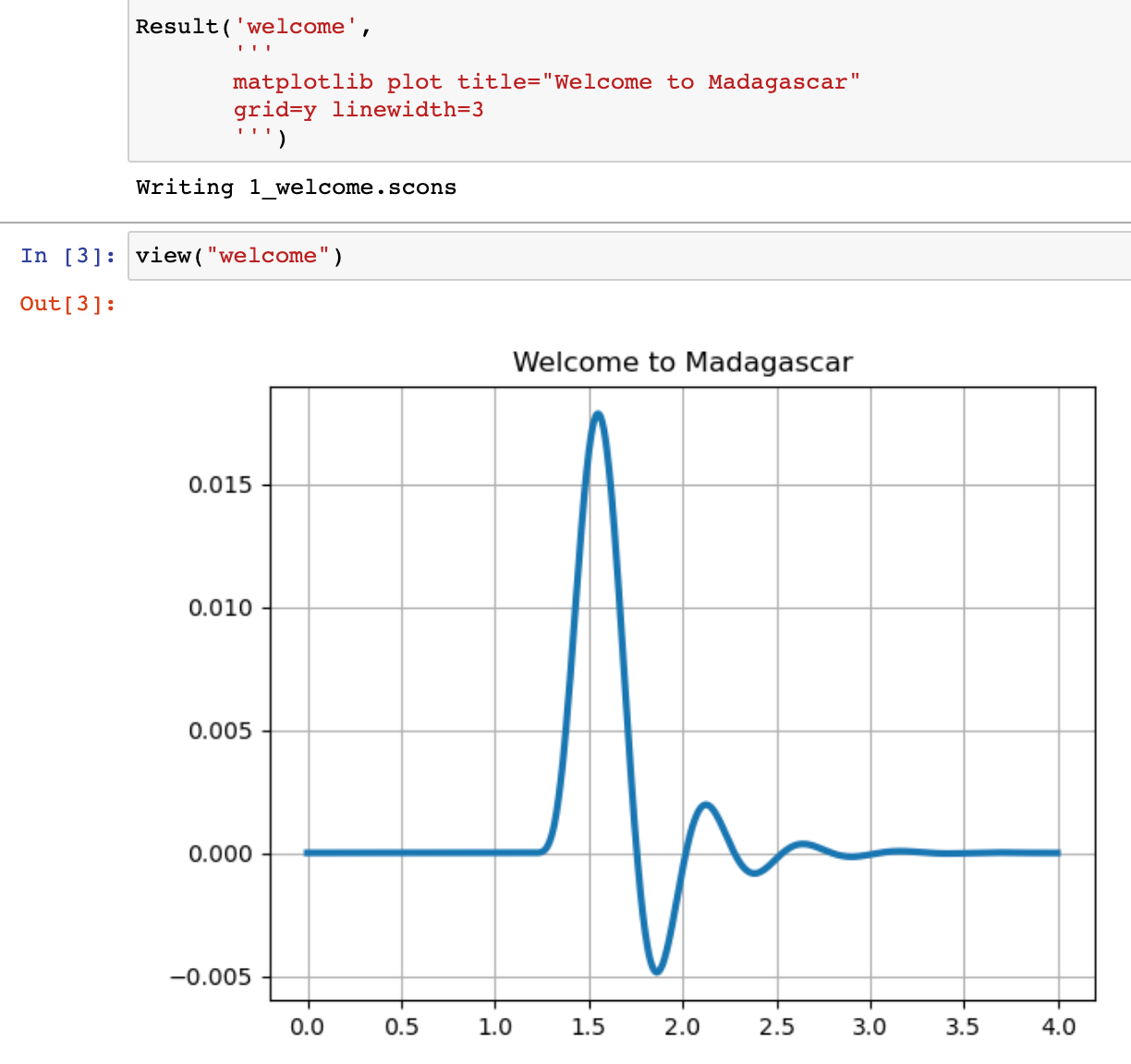
The main advantage of continuing to use Vplot is the availability of sfvplotdiff, a key tool for reproducibility testing and continuous integration.
Madagascar users are invited to try the new functionality and contribute to its further development.





 Is it a good colormap? We can find out by using tools from
Is it a good colormap? We can find out by using tools from 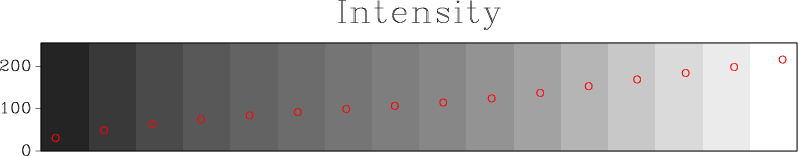
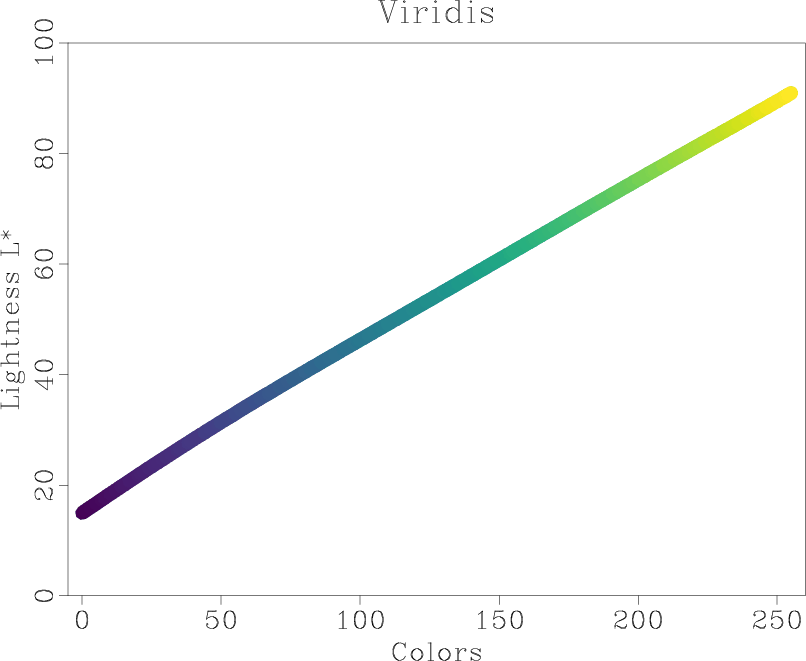
 This analysis shows the intensity and lightness distributions of “viridis” are nicely linear. In his presentation at
This analysis shows the intensity and lightness distributions of “viridis” are nicely linear. In his presentation at