|
|
|
|
Adaptive prediction filtering in |

|
|---|
|
data
Figure 11. 3D field data. |
|
|

|
|---|
|
tpre1
Figure 12. The denoised result by using 3D |
|
|

|
|---|
|
ppred3
Figure 13. The denoised result by using 3D |
|
|
|
|---|
|
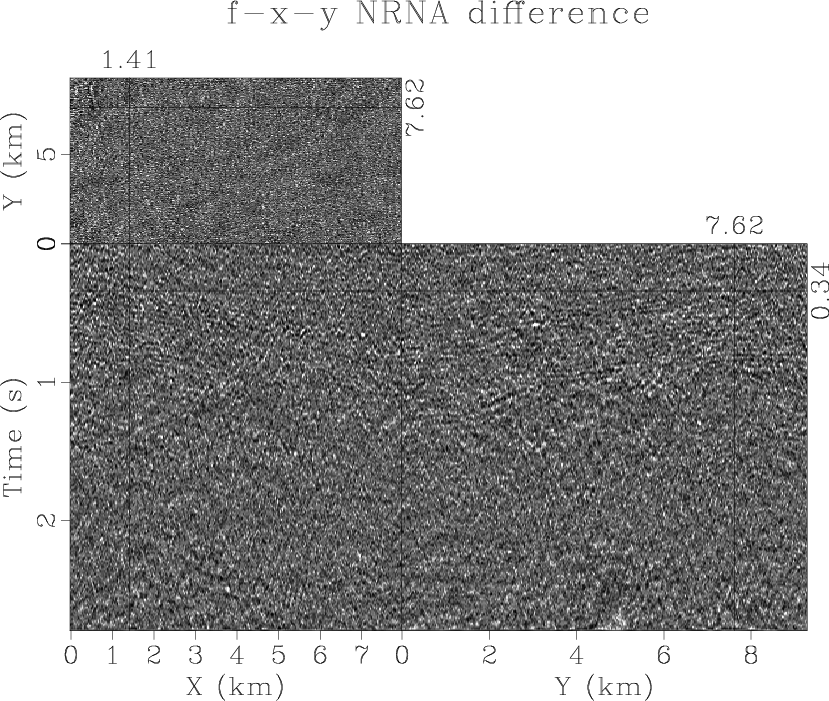
diff3d
Figure 14. The difference between the noisy data (Figure 11) and the denoised result by using 3D |
|
|

|
|---|
|
diff3
Figure 15. The difference between the noisy data (Figure 11) and the denoised result by using 3D |
|
|
|
|
|
|
Adaptive prediction filtering in |