|
|
|
|
Streaming orthogonal prediction filter in |

|
|---|
|
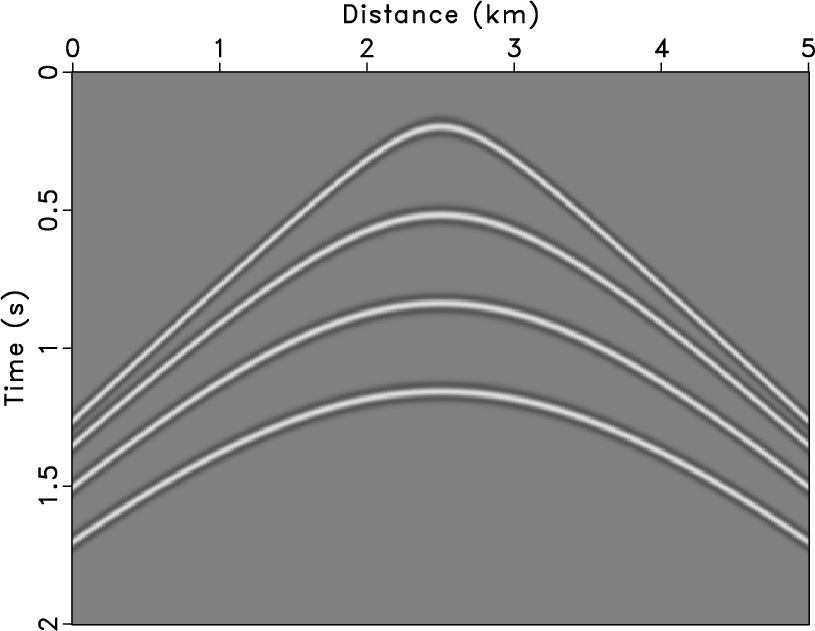
gpara,gnpara
Figure 5. Shot gather (a) and noisy data (b). |
|
|


|
|---|
|
gfx,gfxn2
Figure 6. Denoised result by the |
|
|


|
|---|
|
grna,grnan2
Figure 7. Denoised result by the |
|
|


|
|---|
|
gh2,gr2
Figure 8. Denoised result by the |
|
|


|
|---|
|
diff1,diff2
Figure 9. Comparison of the difference between Figure 5a and the corresponding denoised result using two different methods, the nonstationary |
|
|
|
|
|
|
Streaming orthogonal prediction filter in |